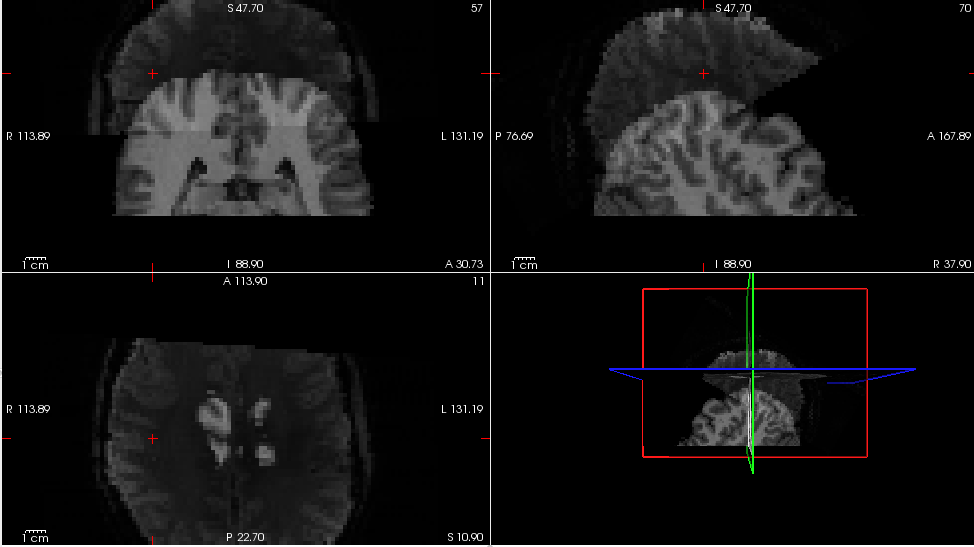
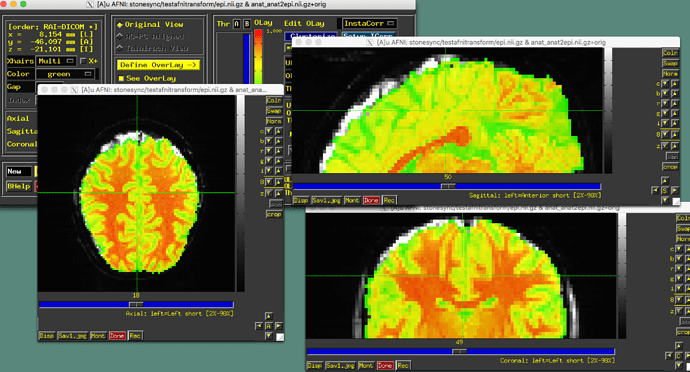
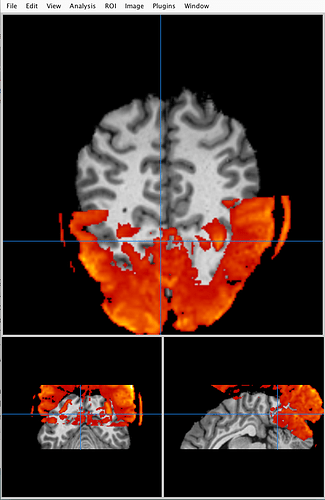
I just recently found that afni, FSLeyes, Freeview and Mango interpret NIFTI files differently. I performed an epi-anat alignment using AFNI and the alignment looks good in AFNI viewer. But if I visualized the aligned NIFTI files in FSLeys, Freeview or Mango, I can clearly see the mismatch. I know that AFNI uses a LPS+ space and other software use a RAS+ space. But clearly they do not just differ in flipping the coordinates.
My question is, how to somehow modify the header information such that the two files aligned in AFNI viewer can also show up being properly aligned in like FSLeyes or Freeview?
This looks like a NIfTI image header problem. The NIfTI specification allows the orientation to be specified in multiple ways, and different programs use different defaults, so it not every field is specified perfectly (or if a field is used that a particular program ignores), the images won’t be displayed consistently.
I have a bit of information about this on my blog; try the general information in http://mvpa.blogspot.com/2015/04/format-conversion-4dfp-to-nifti-plus.html, particularly the first section.The first image in http://mvpa.blogspot.com/2013/04/working-with-nifti-files-in-r-oronifti.html shows some NIfTI headers in MRIcronN, which is my go-to program to quickly check NIfTI header fields. At a guess, your image probably has something odd in the qoffsets and/or affine parameters fields.
We’ve found that setting both s/q form matrices to the same 6-parameters transform matrix usually works well. (except for some corner cases in AFNI if the affine is not aligned with the canonical axes).