Dear fMRIprep experts,
Thanks for providing such great software.
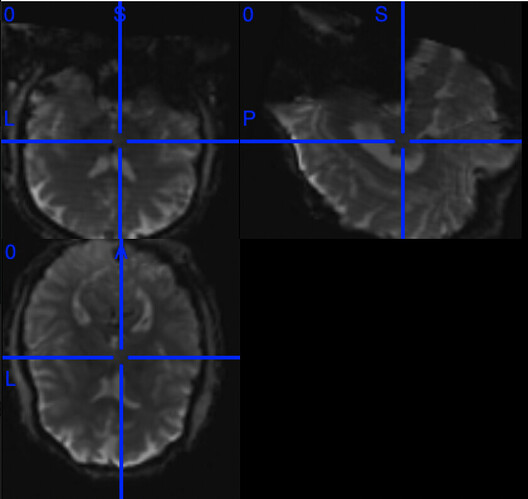
Our lab has used fMRIprep for a while without any issue, however, some recent data collected with GE’s commercial hyperband sequence after a scanner’s upgrade was not processed as expected, that the output from fMRIprep is showing a wrong orientation. For example, below shows the *_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz in Mricron.
My initial check found out the orientation of the new func data was in a different orientation (RPS) than the single-band reference image (LAS) - the first and second axes were rotated. I am wondering if this is causing the issue if fMRIprep assumes that the two images should have the same orientation so co-registration/normalization matrix derived from the single-band reference image is incorrectly applied to the func data.
Orientation info:
func:
| qform_name | Scanner Anat |
|---|---|
| qform_code | 1 |
| qto_xyz:1 | 0.000000 -2.400000 0.000001 102.568001 |
| qto_xyz:2 | -2.335931 0.000000 0.550844 99.948654 |
| qto_xyz:3 | -0.550843 -0.000001 -2.335937 112.630508 |
| qto_xyz:4 | 0.000000 0.000000 0.000000 1.000000 |
| qform_xorient | Anterior-to-Posterior |
| qform_yorient | Right-to-Left |
| qform_zorient | Superior-to-Inferior |
| sform_name | Scanner Anat |
| sform_code | 1 |
| sto_xyz:1 | 0.000000 -2.400000 0.000000 102.568001 |
| sto_xyz:2 | -2.335931 0.000000 0.550842 99.948654 |
| sto_xyz:3 | -0.550843 0.000000 -2.335938 112.630508 |
| sto_xyz:4 | 0.000000 0.000000 0.000000 1.000000 |
| sform_xorient | Anterior-to-Posterior |
| sform_yorient | Right-to-Left |
| sform_zorient | Superior-to-Inferior |
SB reference:
| qform_name | Scanner Anat |
|---|---|
| qform_code | 1 |
| qto_xyz:1 | -2.400000 0.000000 -0.000000 102.568001 |
| qto_xyz:2 | 0.000000 2.335931 -0.550843 -75.449463 |
| qto_xyz:3 | 0.000000 0.550843 2.335931 -74.214508 |
| qto_xyz:4 | 0.000000 0.000000 0.000000 1.000000 |
| qform_xorient | Right-to-Left |
| qform_yorient | Posterior-to-Anterior |
| qform_zorient | Inferior-to-Superior |
| sform_name | Scanner Anat |
| sform_code | 1 |
| sto_xyz:1 | -2.400000 0.000000 -0.000000 102.568001 |
| sto_xyz:2 | -0.000000 2.335931 -0.550844 -75.449463 |
| sto_xyz:3 | 0.000000 0.550843 2.335933 -74.214508 |
| sto_xyz:4 | 0.000000 0.000000 0.000000 1.000000 |
| sform_xorient | Right-to-Left |
| sform_yorient | Posterior-to-Anterior |
| sform_zorient | Inferior-to-Superior |
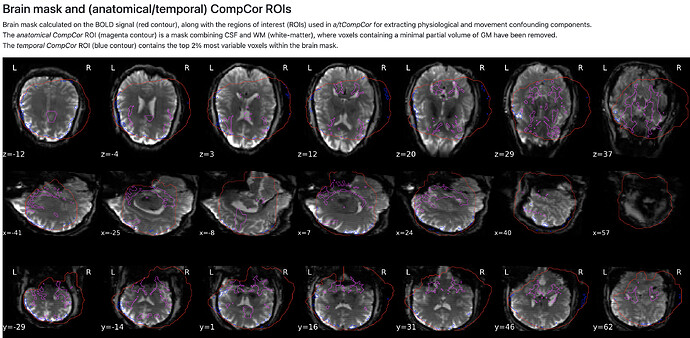
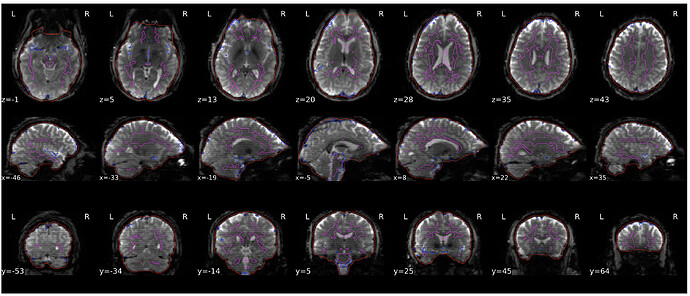
Sorry I can not post the screenshot of the fMRIprep report, because I can only post one screenshot. There was no error or warning, the co-registration looks okay which I think was showing between the SB reference and T1w, however, the CompCorr ROIs and brain mask are showing the wrong underlay with the same orientation issue.
I want to ask:
-
Is there any way to generate the intermediate output from fMRIprep so I can check step by step and figure out what is causing the issue?
-
Currently I am running some tests and expect to have the output in a day, but it would be great to have suggestions from you, I tried:
-
removing SB reference image and re-run fMRIprep
This is not optimal because of the poor contrast of multiband data, just to test if my above guess is correct. -
fslreorient2std func data into the same orientation as the SB reference image.
It seems I will also need to modify the .json file especially the phase encoding direction if I do so, is there any guide on that? I read #260 but am not exactly sure what to modify.
Thanks in advance for your help!
Best,
Xue