Summary of what happened:
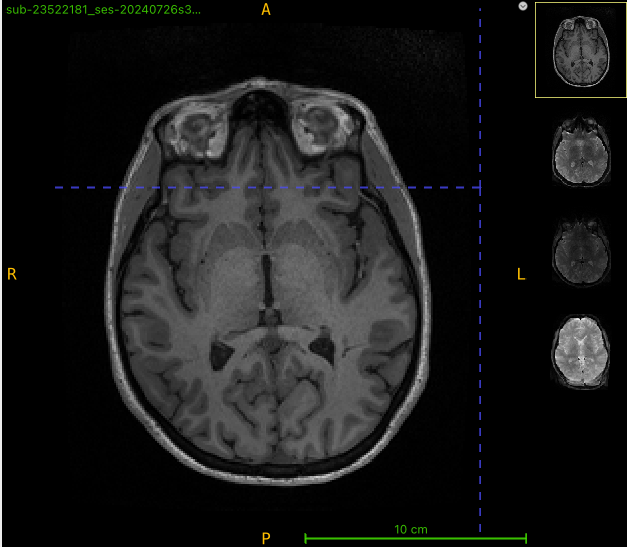
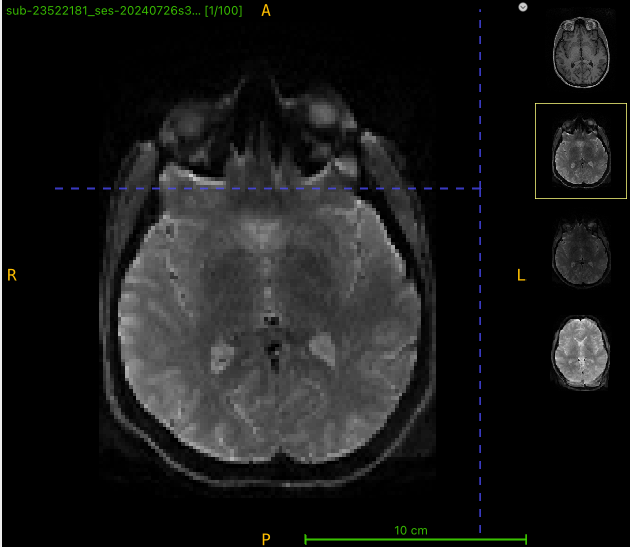
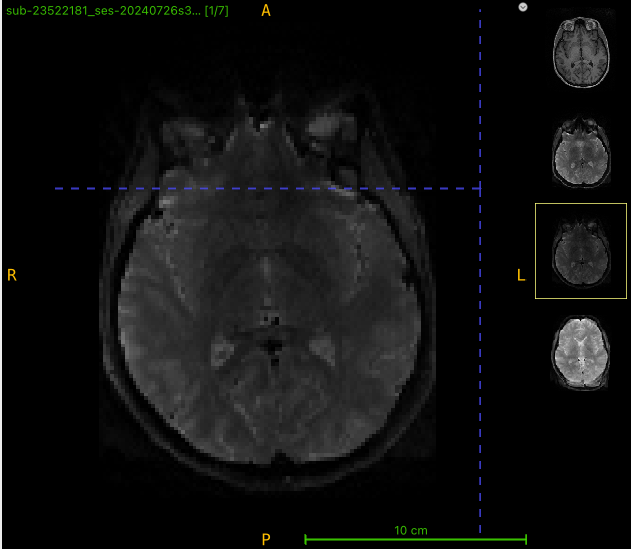
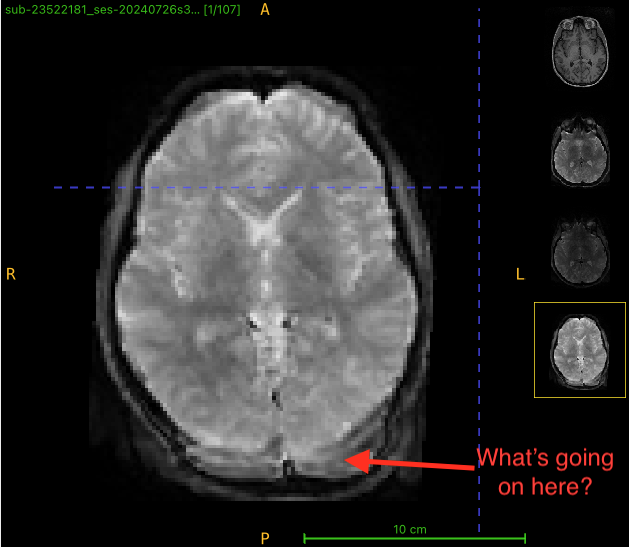
I am getting strange-looking corrected images after running qsiprep. There appears to be extra tissue being created in the output. To see what might be going on, I opened up 4 images (T1w, multi-b dwi, rev phase dwi, and qsiprep output) in ITK-SNAP. The brain shape doesn’t look right for this example, either (looks like a different slice). What is going on?
Command used (and if a helper script was used, a link to the helper script or the command generated):
main script:
#!/bin/bash
qsiprep \
/mnt/share/Bipolar_MDD/Bipolar_MDD_Data /mnt/share/Bipolar_MDD/derivatives/qsiprep/00_preprocessed_dwi participant \
--skip_bids_validation \
--participant-label 23522181 \
--output-resolution 1.7 \
--anat-modality T1w \
--denoise-method dwidenoise \
--unringing-method mrdegibbs \
--b1-biascorrect-stage final \
--distortion-group-merge none \
--anatomical-template MNI152NLin2009cAsym \
--b0-to-t1w-transform Affine \
--intramodal-template-transform Affine \
--fs-license-file /mnt/share/Programs/FreeSurfer7.2/freesurfer/license.txt \
--b0-motion-corr-to iterative \
--hmc-transform Affine \
--hmc_model eddy \
--pepolar-method TOPUP \
--use-syn-sdc \
--verbose \
-w /mnt/share/Bipolar_MDD/derivatives/qsiprep/temp/ \
--bids-filter-file /mnt/share/Bipolar_MDD/scripts/qsiprep//bids_filter_file_sag.json
script for HPC submission:
#!/bin/bash
#$ -pe smp 16
#$ -q PINC,UI
#$ -m bea
#$ -M gail-harmata@uiowa.edu
#$ -j y
#$ -o /Shared/MRRCdata/Bipolar_MDD/scripts/qsiprep//00_preprocessing/out
singularity exec -c -W /nfsscratch/Users/gharmata/ -B /Shared/MRRCdata:/mnt/share /Shared/MRRCdata/Programs/qsiprep/qsiprep-v0.22.1.sif /mnt/share/Bipolar_MDD/scripts/qsiprep//00_preprocessing/job_scripts/sub-23522181_ses-20240726s3T_0.sh
Version:
0.22.2.dev0+g712047b.d20240802
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer/singularity on an HPC
Data formatted according to a validatable standard? Please provide the output of the validator:
There is an issue with our ASL scan naming we need to fix, but that shouldn’t affect qsiprep.
BIDS validator output:
[31m1: [ERR] Files with such naming scheme are not part of BIDS specification. This error is most commonly caused by typos in file names that make them not BIDS compatible. Please consult the specification and make sure your files are named correctly. If this is not a file naming issue (for example when including files not yet covered by the BIDS specification) you should include a ".bidsignore" file in your dataset (see https://github.com/bids-standard/bids-validator#bidsignore for details). Please note that derived (processed) data should be placed in /derivatives folder and source data (such as DICOMS or behavioural logs in proprietary formats) should be placed in the /sourcedata folder. (code: 1 - NOT_INCLUDED)e[39m
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_CBF.json
Evidence: sub-23522181_ses-20240726s3T_CBF.json
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_CBF.nii.gz
Evidence: sub-23522181_ses-20240726s3T_CBF.nii.gz
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_asl.json
Evidence: sub-23522181_ses-20240726s3T_asl.json
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_asl.nii.gz
Evidence: sub-23522181_ses-20240726s3T_asl.nii.gz
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_asla.json
Evidence: sub-23522181_ses-20240726s3T_asla.json
./sub-23522181/ses-20240726s3T/asl/sub-23522181_ses-20240726s3T_asla.nii.gz
Evidence: sub-23522181_ses-20240726s3T_asla.nii.gz
Relevant log outputs (up to 20 lines):
(I don’t see anything relevant in the log)
Screenshots / relevant information:
We collected two sagittal DWI scans: one multi-shell (max b=3000), and a shorter reverse phase encoding (max b=3000, but not a parallel sequence). I tried --distortion-group-merge concat and got an error with that call, so I put --distortion-group-merge none.
Here’s the T1w image:
Here’s the first b0 for the multi-b scan:
Here’s the b0 for the reverse phase scan:
Here’s what qsiprep output (plus annotations):