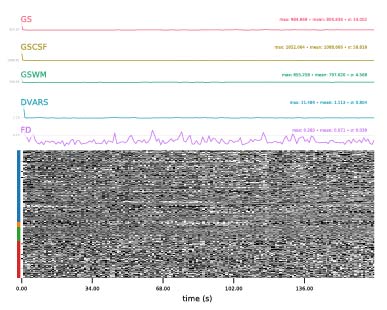
Hi Steven. Thanks very much for the reply–I’ve added mine in turn below. Appreciate your input on this!
Dan
1. What was your command to run fMRIPrep? I’m running this in a singularity container on a cluster (via slurm), and the exact command is “sh fmriprep BIDS fmriprep_output_aroma participant --participant-label REWMEM006 --use-aroma”.
2. What was the error message (can you paste the outputs from the logs)? The error message indicates that ICA-AROMA failed because no signal components were extracted for PST_train run 2 (subjects are doing the probabilistic selection task, aka PST). The logs are long but I’ve copied what I think are the two key parts below:
First this . . .
> [Node] Finished “fmriprep_wf.single_subject_REWMEM006_wf.func_preproc_ses_140127REWMEM006_task_PST_TRAIN_run_2_wf.ica_aroma_wf.ica_aroma”.
211222-05:57:15,484 nipype.workflow INFO:
[Node] Setting-up “fmriprep_wf.single_subject_REWMEM006_wf.func_preproc_ses_140127REWMEM006_task_PST_TRAIN_run_2_wf.ica_aroma_wf.ica_aroma_confound_extraction” in “/net/holynfs01/srv/export/ncf_anl/share_root/anl01/REWMEM/work/fmriprep_wf/single_subject_REWMEM006_wf/func_preproc_ses_140127REWMEM006_task_PST_TRAIN_run_2_wf/ica_aroma_wf/ica_aroma_confound_extraction”.
211222-05:57:15,508 nipype.workflow INFO:
[Node] Running “ica_aroma_confound_extraction” (“fmriprep.interfaces.confounds.ICAConfounds”)
211222-05:57:15,534 nipype.interface WARNING:
No signal components were classified
211222-05:57:15,543 nipype.workflow WARNING:
[Node] Error on “fmriprep_wf.single_subject_REWMEM006_wf.func_preproc_ses_140127REWMEM006_task_PST_TRAIN_run_2_wf.ica_aroma_wf.ica_aroma_confound_extraction” (/net/holynfs01/srv/export/ncf_anl/share_root/anl01/REWMEM/work/fmriprep_wf/single_subject_REWMEM006_wf/func_preproc_ses_140127REWMEM006_task_PST_TRAIN_run_2_wf/ica_aroma_wf/ica_aroma_confound_extraction)
211222-06:12:33,828 nipype.workflow INFO:
. . . and then this, from the very end of the log:
Traceback (most recent call last):
File “/usr/local/miniconda/bin/fmriprep”, line 11, in
sys.exit(main())
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/cli/run.py”, line 342, in main
fmriprep_wf.run(**plugin_settings)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/workflows.py”, line 595, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/base.py”, line 162, in run
self._clean_queue(jobid, graph, result=result))
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/base.py”, line 224, in _clean_queue
raise RuntimeError("".join(result[‘traceback’]))
RuntimeError: Traceback (most recent call last):
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/plugins/multiproc.py”, line 69, in run_node
result[‘result’] = node.run(updatehash=updatehash)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 480, in run
result = self._run_interface(execute=True)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 564, in _run_interface
return self._run_command(execute)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 644, in _run_command
result = self._interface.run(cwd=outdir)
File “/usr/local/miniconda/lib/python3.6/site-packages/nipype/interfaces/base/core.py”, line 521, in run
runtime = self._run_interface(runtime)
File “/usr/local/miniconda/lib/python3.6/site-packages/fmriprep/interfaces/confounds.py”, line 119, in _run_interface
raise RuntimeError(‘ICA-AROMA failed’)
RuntimeError: ICA-AROMA failed
3. Does this persist in the most recent fMRIPrep release? I’m not sure of the answer here yet, as I’m running this on a cluster and I think we only have v1.1.4 installed, which is what I used here. I’ve asked for fmriprep to be updated and when it is I can report back. I think the crashing would certainly stop as my understanding is that that specific problem was fixed, but if nothing else changes I expect that I would still get no signal components for this run and the others that crashed, which is puzzling because they seem fine to me and when I run fmriprep without ICA-AROMA the results look good.
4. Are your data BIDS-valid? Yes, I think so. They’re all in BIDS format and running fmriprep without ICA-AROMA works fine. Also, running fmriprep with ICA-AROMA works fine for 15/22 subjects I have in this dataset.
5. Are the crashes subject-specific or does it fail on everyone? As mentioned above, the crashes are subject-specific: fmriprep with ICA-AROMA works on 15/22 subjects and fails on the other 7, and I’m at a loss for what differentiates the successes from the fails. I collected the data myself on the same scanner in a short span of time, all the participants are community adults, and I don’t see any obvious differences in run quality–in fact, some of the failed runs look cleaner (less motion) than runs that succeeded.
I’ve been trying to think of things to play with in order to gain insight here, and I noticed comments on other threads indicating that global signal regression can sometimes improve ICA-AROMA fits. My understanding is that the global signal (along with CSF and WM covariates) are computed before ICA-AROMA is run. Do you know if there’s a way to ask fmriprep to regress the global signal out of the BOLD data before it’s passed to ICA-AROMA? I thought that might be worth trying, to see if it makes any difference, but I’m not a nipype ninja so am not sure how difficult this would be to do.