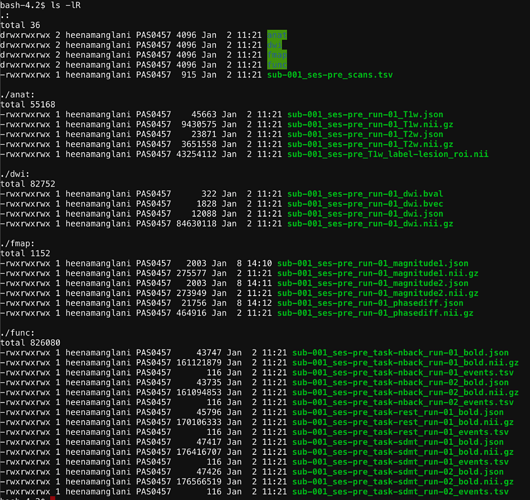
Here is the (long but not verbose) output from running the BIDS validator on the root dir:
bash-4.2$ ./validator_latest.sif /fs/scratch/PAS0457/trac/1_bids/nifti/
bids-validator@1.8.9
bids-specification@disable
(node:5814) Warning: Closing directory handle on garbage collection
(Use node --trace-warnings ... to show where the warning was created)
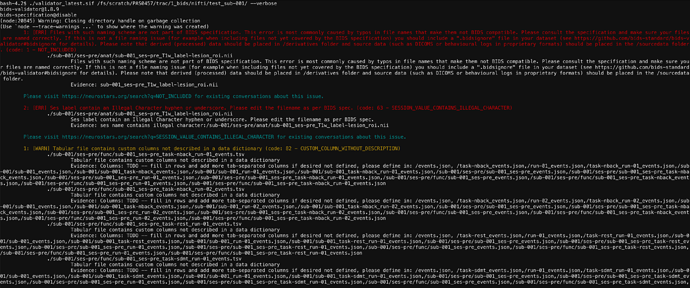
1: [ERR] Invalid JSON file. The file is not formatted according the schema. (code: 55 - JSON_SCHEMA_VALIDATION_ERROR)
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-02_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-02_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-02_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-003/ses-pre/func/sub-003_ses-pre_task-rest_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-003/ses-pre/func/sub-003_ses-pre_task-sdmt_run-01_bold.json
Evidence: .CogAtlasID should match format “uri”
./sub-003/ses-pre/func/sub-003_ses-pre_task-sdmt_run-02_bold.json
Evidence: .CogAtlasID should match format “uri”
… and 328 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=JSON_SCHEMA_VALIDATION_ERROR for existing conversations about this issue.
2: [ERR] Files with such naming scheme are not part of BIDS specification. This error is most commonly caused by typos in file names that make them not BIDS compatible. Please consult the specification and make sure your files are named correctly. If this is not a file naming issue (for example when including files not yet covered by the BIDS specification) you should include a ".bidsignore" file in your dataset (see https://github.com/bids-standard/bids-validator#bidsignore for details). Please note that derived (processed) data should be placed in /derivatives folder and source data (such as DICOMS or behavioural logs in proprietary formats) should be placed in the /sourcedata folder. (code: 1 - NOT_INCLUDED)
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.json
Evidence: sub-031_ses-pre_run-01_T1w.json
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.nii.gz
Evidence: sub-031_ses-pre_run-01_T1w.nii.gz
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.json
Evidence: sub-031_ses-pre_run-01_T2w.json
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.nii.gz
Evidence: sub-031_ses-pre_run-01_T2w.nii.gz
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bval
Evidence: sub-031_ses-pre_run-01_dwi.bval
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bvec
Evidence: sub-031_ses-pre_run-01_dwi.bvec
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.json
Evidence: sub-031_ses-pre_run-01_dwi.json
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.nii.gz
Evidence: sub-031_ses-pre_run-01_dwi.nii.gz
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.json
Evidence: sub-031_ses-pre_run-01_magnitude1.json
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.nii.gz
Evidence: sub-031_ses-pre_run-01_magnitude1.nii.gz
... and 83 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=NOT_INCLUDED for existing conversations about this issue.
3: [ERR] 'IntendedFor' field needs to point to an existing file. (code: 37 - INTENDED_FOR)
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude1.nii.gz
Evidence: func/sub-003_ses-pre_task-nback_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude1.nii.gz
Evidence: func/sub-003_ses-pre_task-nback_run-02_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude1.nii.gz
Evidence: func/sub-003_ses-pre_task-rest_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude1.nii.gz
Evidence: func/sub-003_ses-pre_task-sdmt_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude1.nii.gz
Evidence: func/sub-003_ses-pre_task-sdmt_run-02_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude2.nii.gz
Evidence: func/sub-003_ses-pre_task-nback_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude2.nii.gz
Evidence: func/sub-003_ses-pre_task-nback_run-02_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude2.nii.gz
Evidence: func/sub-003_ses-pre_task-rest_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude2.nii.gz
Evidence: func/sub-003_ses-pre_task-sdmt_run-01_bold.nii.gz
./sub-003/ses-pre/fmap/sub-003_ses-pre_run-01_magnitude2.nii.gz
Evidence: func/sub-003_ses-pre_task-sdmt_run-02_bold.nii.gz
... and 990 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=INTENDED_FOR for existing conversations about this issue.
4: [ERR] Ses label contain an Illegal Character hyphen or underscore. Please edit the filename as per BIDS spec. (code: 63 - SESSION_VALUE_CONTAINS_ILLEGAL_CHARACTER)
./sub-001/ses-pre/anat/sub-001_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-001/ses-pre/anat/sub-001_ses-pre_T1w_label-lesion_roi.nii
./sub-003/ses-pre/anat/sub-003_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-003/ses-pre/anat/sub-003_ses-pre_T1w_label-lesion_roi.nii
./sub-004/ses-pre/anat/sub-004_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-004/ses-pre/anat/sub-004_ses-pre_T1w_label-lesion_roi.nii
./sub-006/ses-pre/anat/sub-006_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-006/ses-pre/anat/sub-006_ses-pre_T1w_label-lesion_roi.nii
./sub-007/ses-pre/anat/sub-007_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-007/ses-pre/anat/sub-007_ses-pre_T1w_label-lesion_roi.nii
./sub-008/ses-pre/anat/sub-008_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-008/ses-pre/anat/sub-008_ses-pre_T1w_label-lesion_roi.nii
./sub-009/ses-pre/anat/sub-009_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-009/ses-pre/anat/sub-009_ses-pre_T1w_label-lesion_roi.nii
./sub-010/ses-pre/anat/sub-010_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-010/ses-pre/anat/sub-010_ses-pre_T1w_label-lesion_roi.nii
./sub-012/ses-pre/anat/sub-012_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-012/ses-pre/anat/sub-012_ses-pre_T1w_label-lesion_roi.nii
./sub-020/ses-pre/anat/sub-020_ses-pre_T1w_label-lesion_roi.nii
Evidence: ses name contains illegal character:/sub-020/ses-pre/anat/sub-020_ses-pre_T1w_label-lesion_roi.nii
... and 58 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=SESSION_VALUE_CONTAINS_ILLEGAL_CHARACTER for existing conversations about this issue.
5: [ERR] Subject label in the filename doesn't match with the path of the file. File seems to be saved in incorrect subject directory. (code: 64 - SUBJECT_LABEL_IN_FILENAME_DOESNOT_MATCH_DIRECTORY)
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.json
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.json is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.nii.gz
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.json
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.json is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.nii.gz
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bval
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bval is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bvec
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bvec is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.json
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.json is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.nii.gz
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.json
Evidence: File: /nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.json is saved in incorrect subject directory as per sub-id in filename.
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.nii.gz
Evidence: File: /nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.nii.gz is saved in incorrect subject directory as per sub-id in filename.
... and 15 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=SUBJECT_LABEL_IN_FILENAME_DOESNOT_MATCH_DIRECTORY for existing conversations about this issue.
6: [ERR] Session label in the filename doesn't match with the path of the file. File seems to be saved in incorrect session directory. (code: 65 - SESSION_LABEL_IN_FILENAME_DOESNOT_MATCH_DIRECTORY)
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.json
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.json is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.nii.gz
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T1w.nii.gz is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.json
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.json is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.nii.gz
Evidence: File: /nback/sub-031/ses-pre/anat/sub-031_ses-pre_run-01_T2w.nii.gz is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bval
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bval is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bvec
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.bvec is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.json
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.json is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.nii.gz
Evidence: File: /nback/sub-031/ses-pre/dwi/sub-031_ses-pre_run-01_dwi.nii.gz is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.json
Evidence: File: /nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.json is saved in incorrect session directory as per ses-id in filename.
./nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.nii.gz
Evidence: File: /nback/sub-031/ses-pre/fmap/sub-031_ses-pre_run-01_magnitude1.nii.gz is saved in incorrect session directory as per ses-id in filename.
... and 14 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=SESSION_LABEL_IN_FILENAME_DOESNOT_MATCH_DIRECTORY for existing conversations about this issue.
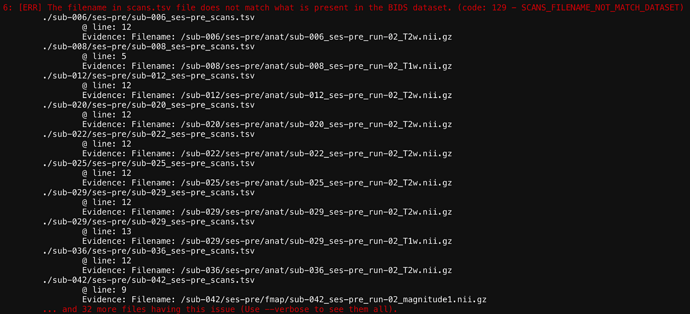
7: [ERR] The filename in scans.tsv file does not match what is present in the BIDS dataset. (code: 129 - SCANS_FILENAME_NOT_MATCH_DATASET)
./sub-006/ses-pre/sub-006_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-006/ses-pre/anat/sub-006_ses-pre_run-02_T2w.nii.gz
./sub-008/ses-pre/sub-008_ses-pre_scans.tsv
@ line: 5
Evidence: Filename: /sub-008/ses-pre/anat/sub-008_ses-pre_run-02_T1w.nii.gz
./sub-012/ses-pre/sub-012_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-012/ses-pre/anat/sub-012_ses-pre_run-02_T2w.nii.gz
./sub-020/ses-pre/sub-020_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-020/ses-pre/anat/sub-020_ses-pre_run-02_T2w.nii.gz
./sub-022/ses-pre/sub-022_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-022/ses-pre/anat/sub-022_ses-pre_run-02_T2w.nii.gz
./sub-025/ses-pre/sub-025_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-025/ses-pre/anat/sub-025_ses-pre_run-02_T2w.nii.gz
./sub-029/ses-pre/sub-029_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-029/ses-pre/anat/sub-029_ses-pre_run-02_T2w.nii.gz
./sub-029/ses-pre/sub-029_ses-pre_scans.tsv
@ line: 13
Evidence: Filename: /sub-029/ses-pre/anat/sub-029_ses-pre_run-02_T1w.nii.gz
./sub-036/ses-pre/sub-036_ses-pre_scans.tsv
@ line: 12
Evidence: Filename: /sub-036/ses-pre/anat/sub-036_ses-pre_run-02_T2w.nii.gz
./sub-042/ses-pre/sub-042_ses-pre_scans.tsv
@ line: 9
Evidence: Filename: /sub-042/ses-pre/fmap/sub-042_ses-pre_run-02_magnitude1.nii.gz
... and 32 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=SCANS_FILENAME_NOT_MATCH_DATASET for existing conversations about this issue.
1: [WARN] Not all subjects contain the same files. Each subject should contain the same number of files with the same naming unless some files are known to be missing. (code: 38 - INCONSISTENT_SUBJECTS)
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-03_bold.json
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-nback_run-03_bold.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-03_bold.nii.gz
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-nback_run-03_bold.nii.gz
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-03_events.tsv
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-nback_run-03_events.tsv
./sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-02_bold.json
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-rest_run-02_bold.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-02_bold.nii.gz
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-rest_run-02_bold.nii.gz
./sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-02_events.tsv
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-rest_run-02_events.tsv
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-04_bold.json
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-sdmt_run-04_bold.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-04_bold.nii.gz
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-sdmt_run-04_bold.nii.gz
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-04_events.tsv
Evidence: Subject: sub-001; Missing file: sub-001_ses-pre_task-sdmt_run-04_events.tsv
./sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-03_bold.json
Evidence: Subject: sub-003; Missing file: sub-003_ses-pre_task-nback_run-03_bold.json
... and 622 more files having this issue (Use --verbose to see them all).
Please visit https://neurostars.org/search?q=INCONSISTENT_SUBJECTS for existing conversations about this issue.
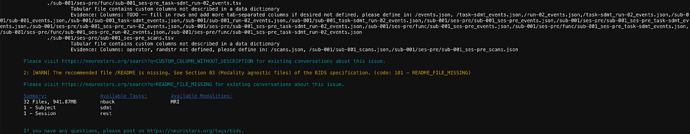
2: [WARN] Tabular file contains custom columns not described in a data dictionary (code: 82 - CUSTOM_COLUMN_WITHOUT_DESCRIPTION)
./participants.tsv
Evidence: Columns: age, sex, group not defined, please define in: /participants.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-01_events.tsv
Evidence: Columns: TODO -- fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-nback_events.json,/run-01_events.json,/task-nback_run-01_events.json,/sub-001/sub-001_events.json,/sub-001/sub-001_task-nback_events.json,/sub-001/sub-001_run-01_events.json,/sub-001/sub-001_task-nback_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-nback_events.json,/sub-001/ses-pre/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-nback_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-01_events.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-02_events.tsv
Evidence: Columns: TODO -- fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-nback_events.json,/run-02_events.json,/task-nback_run-02_events.json,/sub-001/sub-001_events.json,/sub-001/sub-001_task-nback_events.json,/sub-001/sub-001_run-02_events.json,/sub-001/sub-001_task-nback_run-02_events.json,/sub-001/ses-pre/sub-001_ses-pre_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-nback_events.json,/sub-001/ses-pre/sub-001_ses-pre_run-02_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-nback_run-02_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_run-02_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-02_events.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-01_events.tsv
s.json,/sub-001/ses-pre/func/sub-001_ses-pre_run-02_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-02_events.json
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-rest_events.json,/run-01_events.json,/task-rest_run-01_events.json,/sub-0
01/sub-001_events.json,/sub-001/sub-001_task-rest_events.json,/sub-001/sub-001_run-01_events.json,/sub-001/sub-001_task-rest_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-rest_ev
ents.json,/sub-001/ses-pre/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-rest_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-rest_events.json,
/sub-001/ses-pre/func/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-rest_run-01_events.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-01_events.tsv
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-sdmt_events.json,/run-01_events.json,/task-sdmt_run-01_events.json,/sub-0
01/sub-001_events.json,/sub-001/sub-001_task-sdmt_events.json,/sub-001/sub-001_run-01_events.json,/sub-001/sub-001_task-sdmt_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-sdmt_ev
ents.json,/sub-001/ses-pre/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-sdmt_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_events.json,
/sub-001/ses-pre/func/sub-001_ses-pre_run-01_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-01_events.json
./sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-02_events.tsv
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-sdmt_events.json,/run-02_events.json,/task-sdmt_run-02_events.json,/sub-0
01/sub-001_events.json,/sub-001/sub-001_task-sdmt_events.json,/sub-001/sub-001_run-02_events.json,/sub-001/sub-001_task-sdmt_run-02_events.json,/sub-001/ses-pre/sub-001_ses-pre_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-sdmt_ev
ents.json,/sub-001/ses-pre/sub-001_ses-pre_run-02_events.json,/sub-001/ses-pre/sub-001_ses-pre_task-sdmt_run-02_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_events.json,
/sub-001/ses-pre/func/sub-001_ses-pre_run-02_events.json,/sub-001/ses-pre/func/sub-001_ses-pre_task-sdmt_run-02_events.json
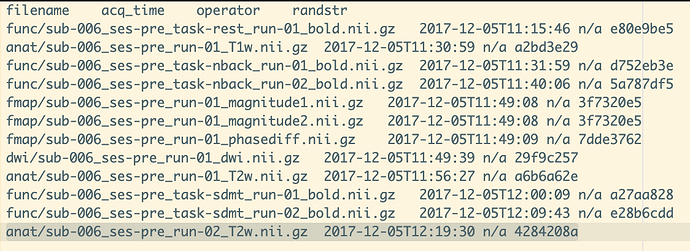
./sub-001/ses-pre/sub-001_ses-pre_scans.tsv
Evidence: Columns: operator, randstr not defined, please define in: /scans.json, /sub-001/sub-001_scans.json,/sub-001/ses-pre/sub-001_ses-pre_scans.json
./sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-01_events.tsv
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-nback_events.json,/run-01_events.json,/task-nback_run-01_events.json,/sub
-003/sub-003_events.json,/sub-003/sub-003_task-nback_events.json,/sub-003/sub-003_run-01_events.json,/sub-003/sub-003_task-nback_run-01_events.json,/sub-003/ses-pre/sub-003_ses-pre_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-nba
ck_events.json,/sub-003/ses-pre/sub-003_ses-pre_run-01_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-nback_run-01_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-nback_event
s.json,/sub-003/ses-pre/func/sub-003_ses-pre_run-01_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-01_events.json
./sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-02_events.tsv
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-nback_events.json,/run-02_events.json,/task-nback_run-02_events.json,/sub
-003/sub-003_events.json,/sub-003/sub-003_task-nback_events.json,/sub-003/sub-003_run-02_events.json,/sub-003/sub-003_task-nback_run-02_events.json,/sub-003/ses-pre/sub-003_ses-pre_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-nba
ck_events.json,/sub-003/ses-pre/sub-003_ses-pre_run-02_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-nback_run-02_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-nback_event
s.json,/sub-003/ses-pre/func/sub-003_ses-pre_run-02_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-nback_run-02_events.json
./sub-003/ses-pre/func/sub-003_ses-pre_task-rest_run-01_events.tsv
Evidence: Columns: TODO – fill in rows and add more tab-separated columns if desired not defined, please define in: /events.json, /task-rest_events.json,/run-01_events.json,/task-rest_run-01_events.json,/sub-0
03/sub-003_events.json,/sub-003/sub-003_task-rest_events.json,/sub-003/sub-003_run-01_events.json,/sub-003/sub-003_task-rest_run-01_events.json,/sub-003/ses-pre/sub-003_ses-pre_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-rest_ev
ents.json,/sub-003/ses-pre/sub-003_ses-pre_run-01_events.json,/sub-003/ses-pre/sub-003_ses-pre_task-rest_run-01_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-rest_events.json,
/sub-003/ses-pre/func/sub-003_ses-pre_run-01_events.json,/sub-003/ses-pre/func/sub-003_ses-pre_task-rest_run-01_events.json
… and 393 more files having this issue (Use --verbose to see them all).
Please visit Search results for 'CUSTOM_COLUMN_WITHOUT_DESCRIPTION' - Neurostars for existing conversations about this issue.
3: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL)
Please visit Search results for 'README_FILE_SMALL' - Neurostars for existing conversations about this issue.
Summary: Available Tasks: Available Modalities:
2120 Files, 63.97GB nback MRI
68 - Subjects sdmt
1 - Session rest
TODO: full task name for nback
TODO: full task name for rest
TODO: full task name for sdmt
I changed the IntendedFor to include the session but still getting the same error:
“IntendedFor”:[
“ses-pre/func/sub-001_ses-pre_task-nback_run-01_bold.nii.gz”,
“ses-pre/func/sub-001_ses-pre_task-nback_run-02_bold.nii.gz”,
“ses-pre/func/sub-001_ses-pre_task-rest_run-01_bold.nii.gz”,
“ses-pre/func/sub-001_ses-pre_task-sdmt_run-01_bold.nii.gz”,
“ses-pre/func/sub-001_ses-pre_task-sdmt_run-02_bold.nii.gz”
],
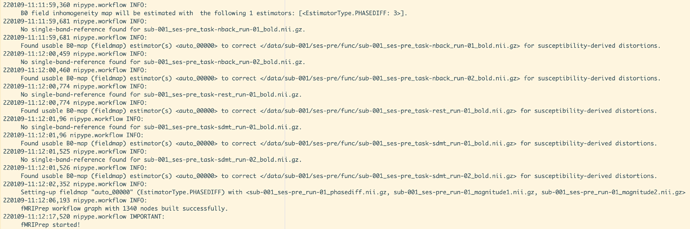
No single-band-reference found for sub-001_ses-pre_task-nback_run-01_bold.nii.gz.
220108-21:14:05,882 nipype.workflow INFO:
Found usable B0-map (fieldmap) estimator(s) <auto_00000> to correct </data/sub-001/ses-pre/func/sub-001_ses-pre_task-nback_run-01_bold.nii.gz> for susceptibility-derived distortions.