Hi all. The official website does not provide the interpretation of the qsiprep output. Could you give me some reference knowledge about this? Thanks!
Hi @bin_liu , are you speaking of the preprocessing outputs or the reconstruction outputs?
Didn’t this page help you?
https://qsiprep.readthedocs.io/en/latest/preprocessing.html#outputs-of-qsiprep
Thanks. This page can tell me information of preprocessing outputs. Could you give me some reference about reconstruction output?
Many of the reconstruction pipelines are coming from specific software, you have to look in the documentation of each of them.
The list of already implemented reconstruction pipelines are here:
https://qsiprep.readthedocs.io/en/latest/reconstruction.html
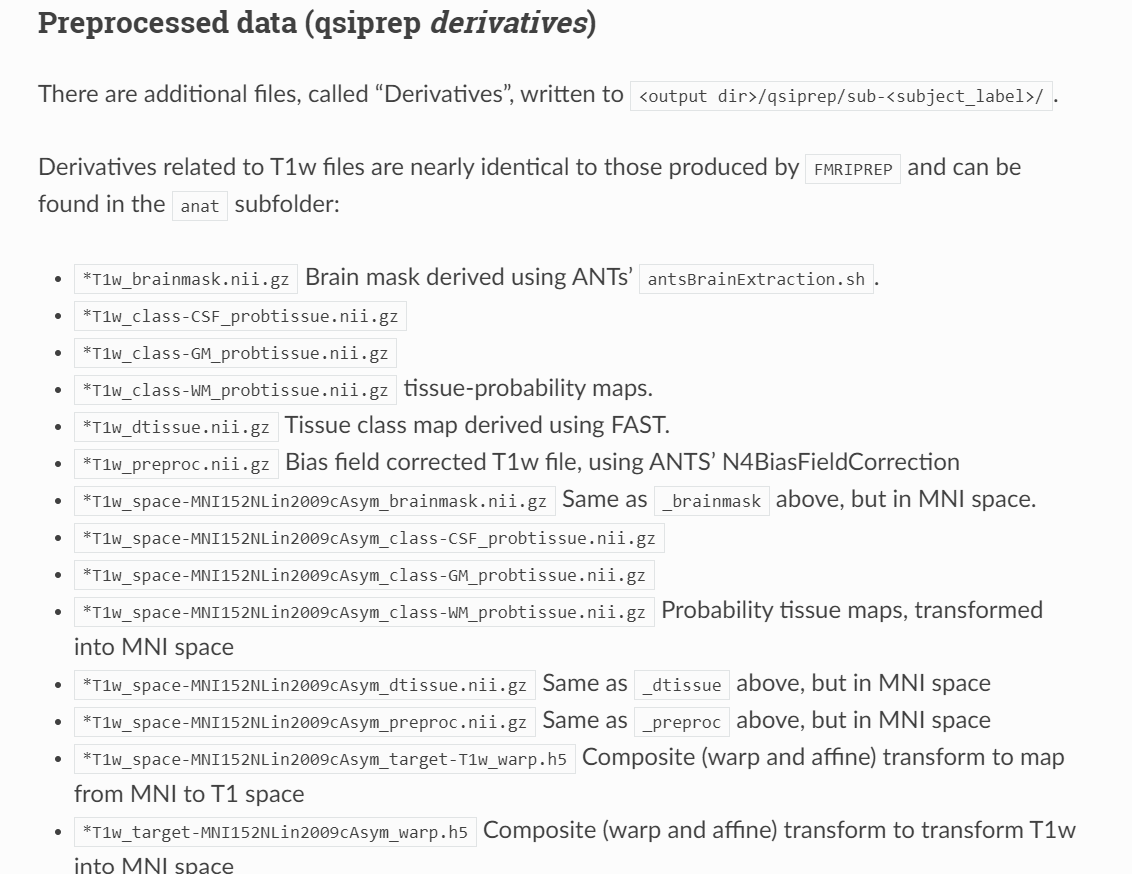
Hi jsein, I looked at the link you share here but it has been moved. I have a similar problem about qsiprep derivatives for T1w in the anat folder.
In the image shared, I cant seem to get the outputs related to “T1w_class-CSF_probtissue.nii.gz`” when i run qsiprep.
It also shows that freesurfer didnt run in the .html file. Can you help me clarify the problem?
Hi @eUgwueke,
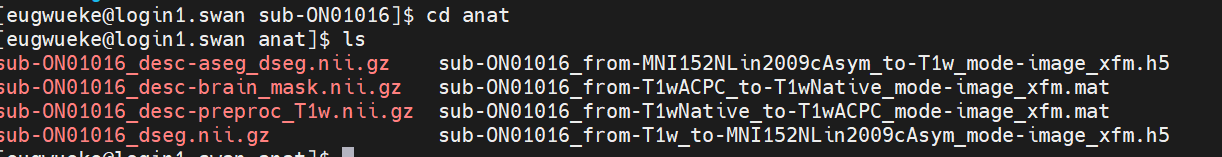
That documentation is not up-to-date. Here is an example of anat outputs in 0.23.0:
sub-XXX_desc-aseg_dseg.nii.gz sub-XXX_from-MNI152NLin2009cAsym_to-T1w_mode-image_xfm.h5
sub-XXX_desc-brain_mask.nii.gz sub-XXX_from-T1wACPC_to-T1wNative_mode-image_xfm.mat
sub-XXX_desc-preproc_T1w.json sub-XXX_from-T1wNative_to-T1wACPC_mode-image_xfm.mat
sub-XXX_desc-preproc_T1w.nii.gz sub-XXX_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5
sub-XXX_dseg.nii.gz
This is not a problem. QSIPrep hasn’t run FreeSurfer for quite a while. recon-all outputs are only used in certain QSIRecon post-processing workflows (those with hsvs 5-tissue-type / 5TT generation).
If you need a CSV mask, it is the 1-valued part of _dseg.nii.gz.
Best,
Steven
I haven’t run 0.19.0 for a while, but if there are no errors, then it should be fine.