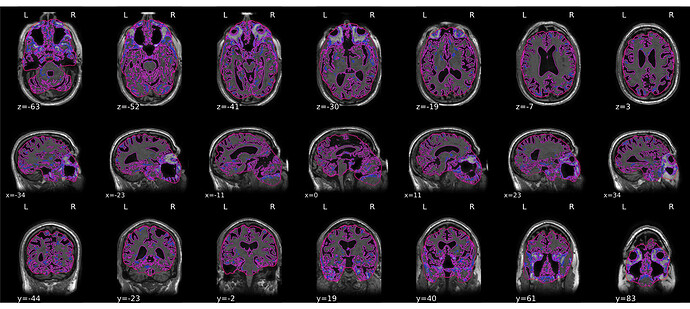
Hello,
After running fmriprep on 10 subjects, 4 of the brain extractions failed and I am unsure of how to troubleshoot this issue (example included). Of note, our anatomical data are PURE corrected. Would that be redundant or cause an issue with the N4BiasField correction?
cmd="${SINGULARITY_CMD}
–participant-label $subject
–ignore fieldmaps slicetiming
–bids-filter-file /data/sen.json
–fs-license-file /data/derivatives/license.txt
–fs-subjects-dir /data/derivatives/T1
–fs-no-reconall
–bold2t1w-dof 6 --nthreads 8 --omp-nthreads 8
–work-dir /work
–write-graph --notrack -v
/data/nifti_test /data/derivatives/FMRI_Sentences participant"
Version 21.0.1
Thank you.