Summary of what happened:
Hi all, I’m using rapidtide version 3.1.4 to extract lag maps for some multi-band resting state data. My cohort has a broad age range (20s-60s) and so I have been toggling with a couple of parameters (search range, smoothing etc). I recently discovered the —delaymapping macro and attempted to call it.
In the documentation, it mentions the following:
“Preset for delay mapping analysis - this is a macro that sets searchrange=(-10.0, 30.0), passes=3, despeckle_passes=4, gausssigma=2.5, refineoffset=True, refinedelay=True, outputlevel=’normal’, dolinfitfilt=False. Any of these options can be overridden with the appropriate additional arguments.”
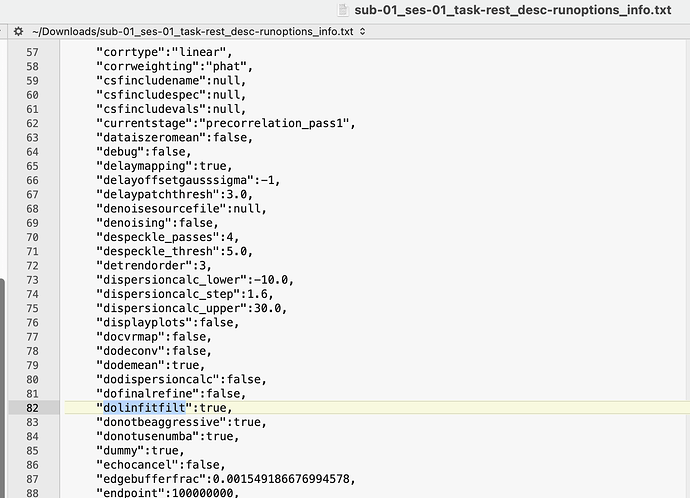
On running a macro test, all —delaymapping parameters seemed to check out except for the ‘dolinfitfilt’ option. Attached is my rapidtide call, and the sub-01_ses-01_task-rest_desc-runoptions_info.json. Inside my json, I noticed that dolinfitfilt was encoded as true instead of false. I would greatly appreciate any suggestions on how to debug and fix this, thank you!
Command used (and if a helper script was used, a link to the helper script or the command generated):
apptainer exec -B /path_to_data/fmriprep/sub-01/ses-01:/data_in \
-B /path_to_data/rapidtide_test_v314_macro_test/sub-01:/data_out \
/path_to_software/sif/rapidtide_latest_v3.1.4.sif rapidtide \
/data_in/func/sub-01_ses-01_task-rest_desc-preproc_bold.nii.gz \
/data_out/sub-01_ses-01_task-rest \
--delaymapping \
--filterband lfo \
--brainmask /data_in/func/sub-01_ses-01_task-rest_desc-brain_mask.nii.gz \
--graymattermask /data_in/fMRIPrep_parc_bold_space/sub-01_ses-01_task-rest_space-bold_desc-GM_probseg_bin.nii.gz \
--whitemattermask /data_in/fMRIPrep_parc_bold_space/sub-01_ses-01_task-rest_space-bold_desc-WM_probseg_bin.nii.gz \
--mklthreads 1 \
--nprocs 1 \
--numskip 0 \
--outputlevel max
Version:
rapidtide v3.1.4
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Relevant log outputs (up to 20 lines):
setting initregressorinclude mask to gray matter mask
setting corrmaskinclude mask to gray matter mask
setting refineinclude mask to gray matter mask
setting offsetinclude mask to gray matter mask
setting initregressorinclude mask to gray matter mask
WARNING:rapidtide.workflows.rapidtide_parser:Using "delaymapping" analysis mode. Overriding any affected arguments.
overriding lagmin
overriding lagmax
overriding dolinfitfilt
pfftw does not exist
no aggressive optimization
will not use numba even if present
garbage collection is on
starting rapidtide v3.1.4
loading data from /data_in/func/sub-01_ses-01_task-rest_desc-preproc_bold.nii.gz
322752 spatial locations, 646 timepoints
startpoint set to 0
endpoint set to maximum, ( 645 )
startpoint set to 0
endpoint set to maximum, ( 645 )
applying gaussian spatial filter to timepoints 0 to 645 with sigma=2.5
Timepoint: 100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 646/646 [00:05<00:00, 126.37timepoints/s]
fmri_data.shape=(322752, 646)
constructing global mean include mask
constructing refine include mask
constructing offset include mask
constructing global mean signal using sum
used 20864 voxels to calculate initial regressor signal
constructing global mean signal using sum
used 60158 voxels to calculate whole brain signal
constructing global mean signal using sum
used 20864 voxels to calculate gray matter signal
constructing global mean signal using sum
used 21232 voxels to calculate white matter signal
constructing global mean signal using sum
used 60158 voxels to calculate CSF signal
garbage collected - unable to collect 543 objects
no regressor file specified - will use the global mean regressor
synctime is 0.0
total probe regressor offset is 0.0
filtering to lfo band
setting up fast resampling with padtime = 59.79999923706055
allocated 330.181 megabytes locally for correlation
allocated 298.100 megabytes locally for delay refinement
allocated 1.391 gigabytes locally for refinement
Screenshots / relevant information:
sub-01_ses-01_task-rest_desc-runoptions_info.txt (10.3 KB)