Summary of what happened:
am currently working on a resting-state fMRI seed-based analysis and have encountered an issue that I hope you can help me resolve. When I overlay my binary ROI mask on the preprocessed fMRIPrep data, the alignment appears accurate without any issues.
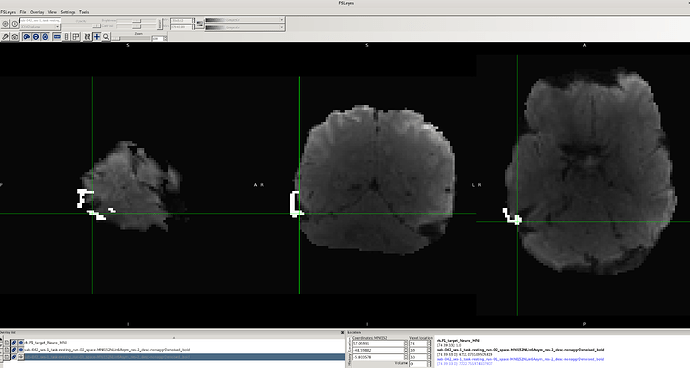
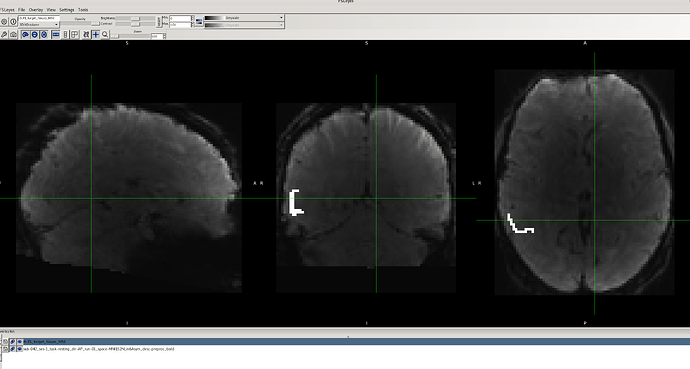
However, after applying the fMRIPost-AROMA processing script, I noticed that for the same data with no issues in the preprocessed fMRIPrep output, the binary ROI mask does not fully align with the denoised BOLD image (output of fMRIPost-AROMA). Additionally, there appears to be distortion in the data for these cases (e.g., sub-042, as shown in the attached images).
It’s worth mentioning that this problem does not occur consistently across all participants. For several participants, there is no misalignment or distortion, either in the preprocessed fMRIPrep or fMRIPost-AROMA processed data. I tried using the flag --output-spaces MNI152NLin6Asym to ensure the output of the fMRIPost-AROMA processing remains in the same standard space, but this flag was not recognized.
I would greatly appreciate your insights into possible reasons for this issue and any suggestions on how to resolve it. Could it be related to the post-processing steps, or is there something else I should investigate in my pipeline?
Thank you in advance for your time and expertise. Please let me know if additional details or data samples would be helpful.
Command used (and if a helper script was used, a link to the helper script or the command generated):
My preprocessing was conducted using fMRIPrep with the following command.
fmriprep_command=(
docker run --rm
-v "$bidsdir:/data"
-v "$outputdir:/out"
-v "$license_file:/usr/local/freesurfer/license.txt"
-v "$workdir:/work"
nipreps/fmriprep:latest
/data /out participant
--participant-label "$pid"
--fs-license-file /usr/local/freesurfer/license.txt
--skip_bids_validation
--no-submm-recon
--n-cpus 16
--omp-nthreads 8
--work-dir /work
--output-spaces MNI152NLin6Asym T1w
)
fmripost-aroma was run with the following command
fMRIPostAROMA_command=(
docker run --rm
-v "$bids_dir:/data"
-v "$output_dir:/data/derivatives"
-v "$ica_outputdir:/out"
-v "$work_dir:/work"
nipreps/fmripost-aroma:latest
/data /out participant
-w /work
--derivatives fmriprep=/data/derivatives
--skip_bids_validation
--participant-label "$pid"
--denoising-method nonaggr
--n-cpus 16
--omp-nthreads 8
)
Version:
PUT VERSION HERE
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE