Summary of what happened:
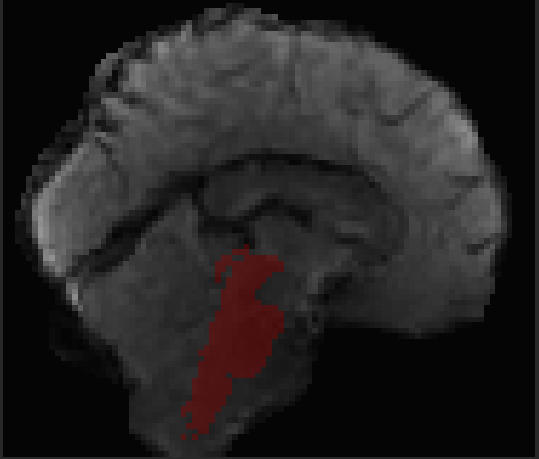
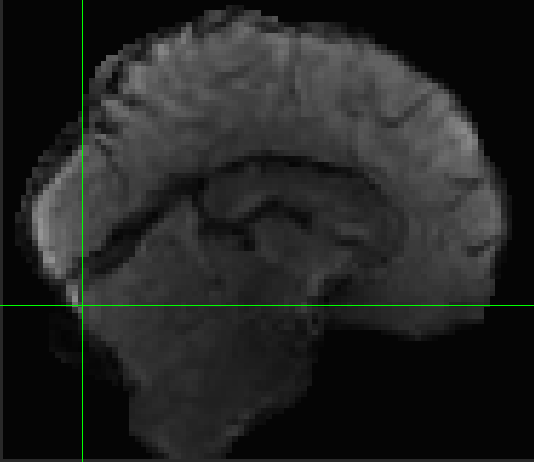
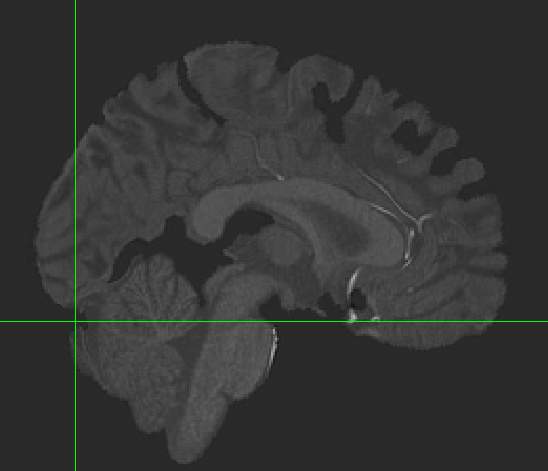
Hi all, I’m hoping to get some suggestions for how to handle alignment in my dataset. I have 3T BOLD images for a set of participants whose structural scans were collected at 7T. When I run fmriprep, the cortex seems to align well between BOLD and T1w images, but everything subcortical looks shifted. The brainstem looks misaligned, and the cerebellum as well. I’ve tried several things and can’t seem to resolve the problem. I’ll post the general code that I’ve used below with a list of different commands I’ve tried.
Things I’ve changed about this:
- I’ve tried running without
use-syn-sdc - I’ve tried adding
--bold2anat-init header - I’ve tried adding
--bold2anat-dof 9
None of these seem to make significant changes in the alignment between my BOLD and T1w data.
Command used (and if a helper script was used, a link to the helper script or the command generated):
while read SUBJ; do
echo "============================="
echo " Running fMRIPrep for $SUBJ "
echo "============================="
# make a subject-specific temp dir
mkdir -p $fmriprep_tmp
singularity run --cleanenv \
-B $bidsdir \
-B $fmriprep_tmp \
-B $cache \
-B $fmriprep_output \
-B ${fs_lic}
$fmriprep \
$bidsdir \
$fmriprep_output \
participant \
--participant-label $SUBJ \
--task-id rest \
--work-dir $fmriprep_tmp \
--output-spaces T1w MNI152NLin2009cAsym \
--ignore slicetiming \
--use-syn-sdc \
--fs-license-file /opt/freesurfer/license.txt \
--derivatives $anat \
--skull-strip-t1w skip \
--fs-subjects-dir $freesurfer \
--fs-no-reconall \
--skip-bids-validation
echo "Finished $SUBJ"
done < "$subj_list"
echo "All subjects processed."
Version:
fmriprep-25.2.3
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
BIDS validated dataset -- no issues that would affect preprocessing
Relevant log outputs (up to 20 lines):
No errors to report