Summary of What Happened
Hello everyone,
I have been facing issues with running QSIPrep on DWI data collected from a Philips 3T scanner, and I am hoping for some guidance to resolve it. I have tried processing both concatenated and non-concatenated DWI files, but I encounter issues with the output structure, including missing HTML reports and what seems like an infinite loop during processing. Additionally, the BIDS validator outputs some warnings that might be relevant.
Command
1. Non-concatenated DWI files (Without Susceptibility Distortion Correction):
For the first attempt, I ran QSIPrep with two separate DWI files and disabled susceptibility distortion correction using the --ignore-topup flag. Here’s the command I used:
singularity run --bind /path/to/license.txt:/opt/freesurfer/license.txt \
--cleanenv /path/to/qsiprep.sif \
/path/to/data /path/to/output \
participant \
--skip_bids_validation \
--separate-all-dwis \
--ignore-topup \
--output-space T1w \
--n_cpus 2 \
--omp-nthreads 2 \
--mem_mb 16000 \
-w /path/to/work \
--verbose \
--output-resolution 1.5
2. Concatenated DWI file (Also Without Susceptibility Distortion Correction):
I also tried concatenating the two DWI scans into a single file and re-ran QSIPrep without susceptibility distortion correction (--ignore-topup), this time using Apptainer:
apptainer run --cleanenv \
-B /path/to/data:/data:ro \
-B /path/to/output:/out \
-B /path/to/license.txt:/opt/freesurfer/license.txt \
/path/to/qsiprep.sif \
/data /out participant \
--fs-license-file /opt/freesurfer/license.txt \
--output-resolution 1.5 \
--n_cpus 4 \
--omp-nthreads 4 \
--mem_mb 8000 \
--ignore fieldmaps
Version
- QSIPrep version: v0.23.1.dev0+g634483f.d20240830
- Apptainer version: 1.3.0
Environment
I am running QSIPrep using Singularity/Apptainer containers.
Data Format and BIDS Validation Output
The data structure includes:
/data/sub-XX/ses-01/anat– T1-weighted anatomical images./data/sub-XX/ses-01/dwi– Contains the two separate DWI runs (for the non-concatenated attempt) or the concatenated DWI file./data/sub-XX/ses-01/fmap– Includes AP and PA fieldmaps withIntendedForfields pointing to the DWI file./data/sub-XX/ses-01/func– Functional MRI files.
In the fieldmap JSON files, I have specified the IntendedFor field as follows:
- AP fieldmap JSON:
"IntendedFor": "ses-01/dwi/sub-22_ses-01_dir-AP_dwi.nii.gz" - PA fieldmap JSON:
"IntendedFor": "ses-01/dwi/sub-22_ses-01_dir-AP_dwi.nii.gz"
I ran the BIDS validator and received the following warnings:
1: [WARN] You should define 'SliceTiming' for this file. If you don't provide this information slice time correction will not be possible.
./sub-22/ses-01/func/sub-22_ses-01_task-rest_run-01_bold.nii.gz
./sub-22/ses-01/func/sub-22_ses-01_task-rest_run-02_bold.nii.gz
2: [WARN] Not all subjects/sessions/runs have the same scanning parameters.
./sub-22/ses-01/func/sub-22_ses-01_task-rest_run-02_bold.nii.gz
Problem Description and Observations
1. Loop or No Output when Running Without Susceptibility Distortion Correction:
In the run with non-concatenated DWI files, with susceptibility distortion correction disabled (--ignore-topup), the QSIPrep process seems to enter an infinite loop and does not produce a final HTML report. The only HTML outputs generated are in the figures folder, which lack comprehensive information on processing errors or completed steps.
2. Error with Susceptibility Distortion Correction Enabled (topup):
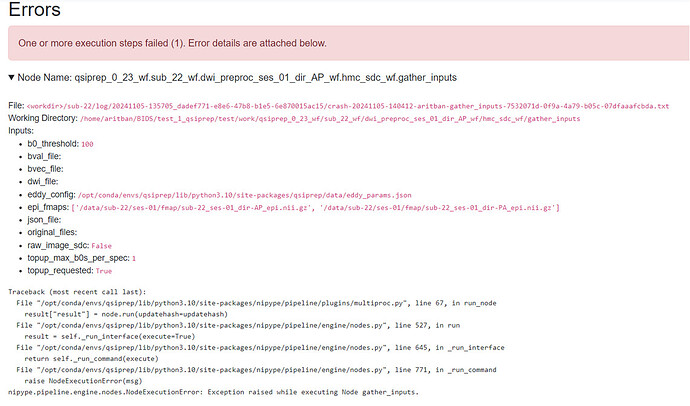
When I enabled topup for susceptibility distortion correction and used the concatenated DWI file, the pipeline produced an HTML report but returned an error in the node gather_inputs. Here’s the error message (screenshot attached):
- Node Name:
qsiprep_0_23_wf.sub_22_wf.dwi_preproc_ses_01_dir_AP_wf.hmc_sdc_wf.gather_inputs - Error:
- The inputs listed include paths to
eddy_params.jsonandepi_fmaps, which appear to be correctly specified. However, the process fails, potentially due to a configuration or compatibility issue.
3. BIDS Validation Warnings:
The BIDS validator outputs warnings about missing SliceTiming information in the functional files, which could affect slice timing correction, and about inconsistent parameters across runs. I’m uncertain if these issues are contributing to the QSIPrep processing problems.
Relevant Log Outputs
Here is a portion of the error traceback when running with the concatenated DWI file and topup (susceptibility distortion correction) enabled:
Traceback (most recent call last):
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/qsiprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node gather_inputs.
Screenshot / Additional Information
Please see the attached screenshot for more details on the error message.
Questions:
- Has anyone experienced similar issues with QSIPrep on Philips 3T data, particularly with susceptibility distortion correction?
- Could the BIDS validation warnings (e.g., missing SliceTiming information and inconsistent parameters) be contributing to the QSIPrep issues?
- Are there specific QSIPrep parameters or setup adjustments that might help resolve the looping issue or help generate the missing HTML report?
Any insights or suggestions to troubleshoot this would be greatly appreciated. Please let me know if you have any additional questions. Thank you so much!