Hello,
I am using netpyne to construct a biophysically realistic cortico-striatal network, and the neuron model type is Hodgkin–Huxley model. Since the Local field potential is electric potential recorded in the extracellular space in brain tissue, it normally fluctuates around baseline 0, at least that’s what we observed from animal experiments. However, the LFP signals recorded from my simulated network consistently display a clear baseline offset, fluctuating below 0. This offset becomes more negative as the simulated electrode is placed deeper. Here is one of the striatal LFP channels I recorded:
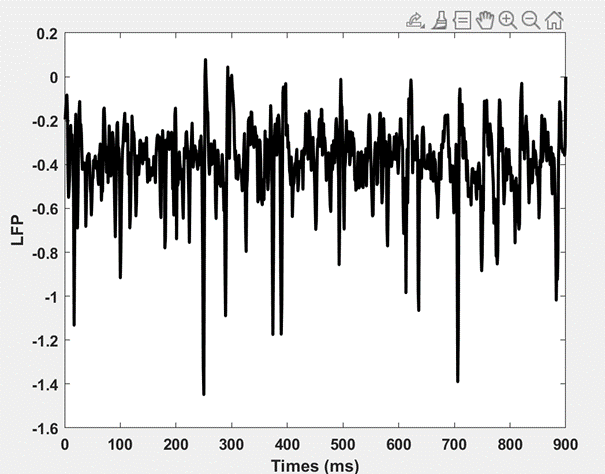
When I divide this cortico-striatal network into two segments, the cortex, and striatum, and simulate them separately, both segments exhibit normal LFP traces that fluctuate around 0. However, as soon as I connect them with synaptic connections, their LFP signals start shifting away from the baseline, primarily below it. While there are numerous preprocessing tools like detrending that can potentially address this issue, I am eager to identify the precise cause.
I would greatly appreciate any guidance on what I might be doing incorrectly or how I could troubleshoot this issue.
Thanks a lot in advance!