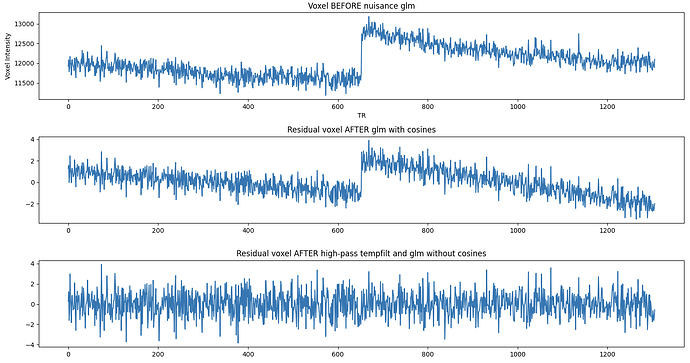
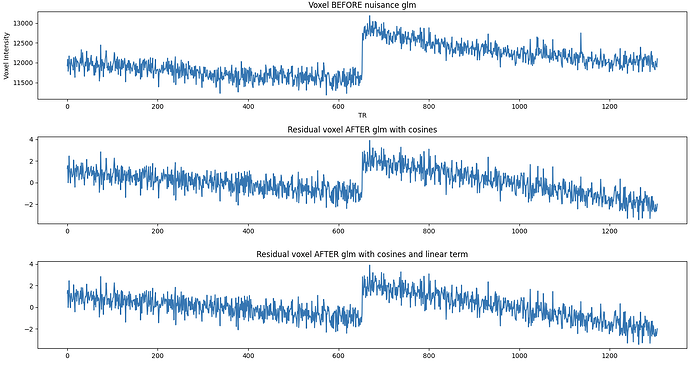
Hi, I’m working with resting-state fMRI data acquired at 7 Tesla, preprocessed with fMRIPrep. Random linear signal drifts (both increasing and decreasing) seem to be present in some, but not all, voxels. What is surprising is that these linear drifts persist after nuisance regression (glm) including discrete cosine functions estimated by fMRIPrep. Any ideas on why these are not being picked up and removed by the model? A temporal high-pass filter (1/128Hz) does seem to remove them adequately, but I’m a bit hesitant to apply temporal filtering before or after the glm given that this can potentially re-introduce artifacts (e.g., Modular preprocessing pipelines can reintroduce artifacts into fMRI data - PubMed). Also, I’m not sure how the DCT regression would behave on already temporally high-pass filtered data? To circumvent this issue, I’ve tried adding a linear term to the glm, which seems very ineffective - which is also surprising to me.
Below, I pasted the regressors in the glm design matrix and plots that show the drifts before after the different steps (two BOLD runs of ~640 volumes each, glm is run separately per run but I’ve concatenated them for plotting). I appreciate any tips or advise!
regressors = [list(np.linspace(-1, 1, len(confounds))), list(confounds.csf), list(confounds.white_matter), list(confounds.framewise_displacement), list(confounds.dvars), list(confounds.rot_x), list(confounds.rot_y), list(confounds.rot_z), list(confounds.trans_x), list(confounds.trans_y), list(confounds.trans_z)] + [list(confounds[col]) for col in confounds.columns if 'cosine' in col] + [list(confounds[col]) for col in confounds[['a_comp_cor_' + str(x).zfill(2) for x in range(21)]]]