Summary of what happened:
I am attempting to use nilearn.interfaces.fmriprep.load_confounds to apply global signal regression (GSR) to my data. My data was processed in fmriprep using AROMA. Here is the exact app that I used to process the data: app-fmriprep. My data was processed on Brainlife and then saved locally, so my files are no longer named according to the fmriprep conventions, but I did my best to BIDS-ify them.
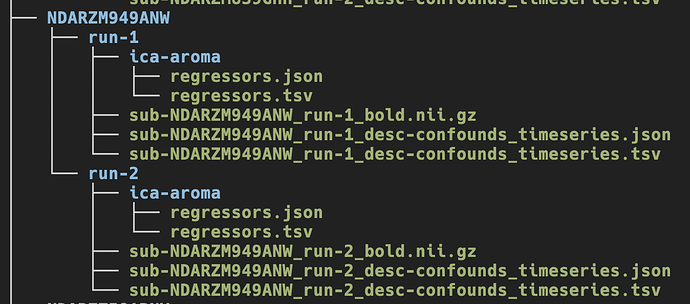
This is an example of what the data looks like:
The ica-aroma files are in their own subfolder because I am not using them currently. Or at least, I am trying not to use them currently.
Command used (and if a helper script was used, a link to the helper script or the command generated):
The rest of my code processes and extracts values from the timeseries. Without this section, the code runs just fine.
confounds_gsr, mask_gsr = load_confounds(
cleaned_epi_path,
strategy=["global_signal"],
global_signal="basic")
cleaned_epi_path is the path to the bold file.
Version:
Python 3.10.9. Nilearn 0.10.0.
Environment (Docker, Singularity, custom installation):
Locally on Ubuntu server
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "removed-this-string-for-security-reasons.py", line 21, in <module>
confounds_gsr, mask_gsr = load_confounds(
File "/home/evb32/.local/lib/python3.10/site-packages/nilearn/interfaces/fmriprep/load_confounds.py", line 275, in load_confounds
sample_mask, conf = _load_confounds_for_single_image_file(
File "/home/evb32/.local/lib/python3.10/site-packages/nilearn/interfaces/fmriprep/load_confounds.py", line 304, in _load_confounds_for_single_image_file
confounds_file = _get_confounds_file(image_file,
File "/home/evb32/.local/lib/python3.10/site-packages/nilearn/interfaces/fmriprep/load_confounds_utils.py", line 121, in _get_confounds_file
_check_images(image_file, flag_full_aroma)
File "/home/evb32/.local/lib/python3.10/site-packages/nilearn/interfaces/fmriprep/load_confounds_utils.py", line 207, in _check_images
raise ValueError(error_message)
ValueError: Invalid file type for the selected method.
I can see where the error is coming from (here) but I can’t figure out what is wrong with my data. The confounds files seem to be formatted correctly. The bold files look normal.
Screenshots / relevant information:
Things I have tried that did not work:
- adding aroma to the pipeline
- renaming the bold file to contain
desc-smoothAROMAnonaggr_bold - renaming the ica-aroma folder
- renaming the timeseries files from
regressors.tsvandregressors.jsonto their current names - trying every available option for
global_signal(“basic”, “full”, etc)