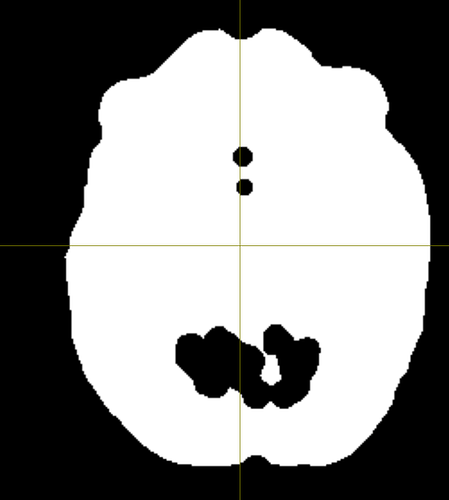
Summary of what happened:
I have been running the MCRIBS pipeline as a docker to look at the segmentations it produces on some neonatal T2 data. *I have had issues running the source code hence why I have used the docker
Whilst the pipeline has produced a good segmentation for one subject, for quite a few of the others I have been facing a problem; specifically, the brain_mask generated from post-processing of the initial bet_mask contains quite a few holes, which is causing some issues later on in the pipeline during surface inflation.
I wanted to ask if anyone else had experienced this issue before and if so, if there was any way to correct the post-processing of the bet mask to stop this issue, or alternatively if there was a way to force the pipeline to select the initial BET mask instead of the post-processed one, as the initial BET mask produced does not contain these holes.
Command used (and if a helper script was used, a link to the helper script or the command generated):
sudo docker run --rm -it --mount type=bind,source=/PATH_TO_DATA,target=/work --mount type=bind,source=PATH_TO_LICENSE,target=/opt/freesurfer-license.txt developmentalimagingmcri/mcribs:latest MCRIBReconAll --all SUBJECT_ID
Version:
docker pull developmentalimagingmcri/mcribs:latest
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE