Summary of what happened:
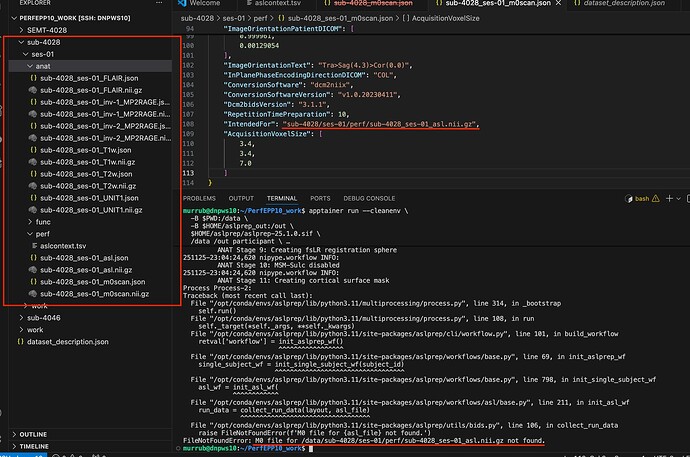
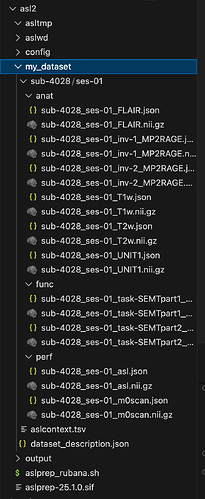
Hi, I posted here a while ago talking about how I kept receiving ‘desc’ image issues, I’ve since fixed that by redownloading the asl apptainer image in my folder. The current issue I’m getting is FileNotFoundError: M0 file for /data/raulab/asl2/my_dataset/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz not found.. I’ve had this issue at the start, and I could fix it with your help by fixing my intededfor path to: "ses-01/perf/sub-4028_ses-01_m0scan.nii.gz". However, I’m still getting the same issue. I’m very confused as to how to move forward from this/why it might be happening, when I pass my dataset through the bids validator I don’t get any red banner errors, so things/my M0 file seems fine there. I would really appreciate any insight/help on this - am very confused. Thank you!!
Command used (and if a helper script was used, a link to the helper script or the command generated): submitting aslprep thourgh a slurm script, pasted below
#!/bin/bash
#SBATCH --job-name=ASLprep_sub4028
#SBATCH --output=/data/raulab/asl2/output/logs/aslprep_%j.out
#SBATCH --error=/data/raulab/asl2/output/logs/aslprep_%j.err
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=16
#SBATCH --time=10:00:00
#SBATCH --mail-user=rubana.mursh@gmail.com
#SBATCH --mail-type=FAIL
## cd /data/raulab/asl2/output/logs
## sbatch /data/raulab/asl2/aslprep_rubana.sh
# Environment Variables
export FS_LICENSE=/data/raulab/asl2/config/license.txt
export TEMPLATEFLOW_HOME=/data/raulab/asl2/config/templateflow
export APPTAINERENV_FS_LICENSE=/config/license.txt
export APPTAINERENV_TEMPLATEFLOW_HOME=/templateflow
#export TMPDIR=/data/raulab/asl2/asltmp
# Node-local work directory
WORKDIR=/data/raulab/asl2/aslwd
echo "Using workdir: ${WORKDIR}"
apptainer run --cleanenv \
--env TEMPLATEFLOW_HOME=/templateflow \
--bind /data/raulab/asl2/my_dataset:/bids:ro \
--bind /data/raulab/asl2/output:/out \
--bind /data/raulab/asl2/config:/freesurfer_license \
--bind /data/raulab/asl2/config/templateflow:/templateflow \
--bind ${WORKDIR}:/work \
/data/raulab/asl2/aslprep-25.1.0.sif \
/bids /out participant \
--participant-label 4028\
--skip_bids_validation \
--output-spaces T1w MNI152NLin2009aSym \
--random-seed 42 \
--n_cpus 16 \
--basil \
--skull-strip-fixed-seed \
--fs-license-file /freesurfer_license/license.txt \
--work-dir /work \
--clean-workdir
Version:
25.1.0
Environment (Docker, Singularity / Apptainer, custom installation):
Apptainer
Data formatted according to a validatable standard? Please provide the output of the validator:
{
"issues": {
"issues": [
{
"code": "README_FILE_MISSING",
"severity": "warning",
"location": "/dataset_description.json",
"rule": "rules.checks.hints.ReadmeFileMissing"
},
{
"code": "TOO_FEW_AUTHORS",
"severity": "warning",
"location": "/dataset_description.json",
"rule": "rules.checks.hints.TooFewAuthors"
},
{
"code": "JSON_KEY_RECOMMENDED",
"subCode": "License",
"severity": "warning",
"location": "/dataset_description.json",
"issueMessage": "Field description: The license for the dataset.\nThe use of license name abbreviations is RECOMMENDED for specifying a license\n(see [Licenses](SPEC_ROOT/appendices/licenses.md)).\nThe corresponding full license text MAY be specified in an additional\n`LICENSE` file.\n",
"rule": "rules.dataset_metadata.dataset_description"
},
{
"code": "JSON_KEY_RECOMMENDED",
"subCode": "GeneratedBy",
"severity": "warning",
"location": "/dataset_description.json",
"issueMessage": "Field description: Used to specify provenance of the dataset.\n",
"rule": "rules.dataset_metadata.dataset_description"
},
{
"code": "JSON_KEY_RECOMMENDED",
"subCode": "SourceDatasets",
"severity": "warning",
"location": "/dataset_description.json",
"issueMessage": "Field description: Used to specify the locations and relevant attributes of all source datasets.\nValid keys in each object include `\"URL\"`, `\"DOI\"` (see\n[URI](SPEC_ROOT/common-principles.md#uniform-resource-indicator)), and\n`\"Version\"` with\n[string](https://www.w3schools.com/js/js_json_datatypes.asp)\nvalues.\n",
"rule": "rules.dataset_metadata.dataset_description"
},
{
"code": "NO_AUTHORS",
"subCode": "Authors",
"severity": "warning",
"location": "/dataset_description.json",
"issueMessage": "Field description: List of individuals who contributed to the creation/curation of the dataset.\n",
"rule": "rules.dataset_metadata.dataset_authors"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-1_MP2RAGE.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-1_MP2RAGE.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-1_MP2RAGE.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "MatrixCoilMode",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T2w.nii.gz",
"issueMessage": "Field description: (If used)\nA method for reducing the number of independent channels by combining in\nanalog the signals from multiple coil elements.\nThere are typically different default modes when using un-accelerated or\naccelerated (for example, `\"GRAPPA\"`, `\"SENSE\"`) imaging.\n",
"rule": "rules.sidecars.mri.MRIHardware"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T2w.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T2w.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T2w.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T1w.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T1w.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_T1w.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_FLAIR.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_FLAIR.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_FLAIR.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-2_MP2RAGE.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-2_MP2RAGE.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_inv-2_MP2RAGE.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_UNIT1.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "SpoilingType",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_UNIT1.nii.gz",
"issueMessage": "Field description: Specifies which spoiling method(s) are used by a spoiled sequence.\n",
"rule": "rules.sidecars.mri.SpoilingType"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_UNIT1.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "FlipAngle",
"severity": "warning",
"location": "/sub-4028/ses-01/anat/sub-4028_ses-01_UNIT1.nii.gz",
"issueMessage": "Field description: Flip angle (FA) for the acquisition, specified in degrees.\nCorresponds to: [DICOM Tag 0018, 1314](https://dicomlookup.com/dicomtags/(0018,1314)) `Flip Angle`.\nThe data type number may apply to files from any MRI modality concerned with\na single value for this field, or to the files in a\n[file collection](SPEC_ROOT/appendices/file-collections.md)\nwhere the value of this field is iterated using the\n[`flip` entity](SPEC_ROOT/appendices/entities.md#flip).\n",
"rule": "rules.sidecars.mri.MRIFlipAngleLookLockerFalse"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_m0scan.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_m0scan.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "TOTAL_ACQUIRED_VOLUMES_NOT_CONSISTENT",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"rule": "rules.checks.asl.ASLTotalAcquiredPairsASLContextLength"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "VascularCrushing",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Boolean indicating if Vascular Crushing is used.\nCorresponds to [DICOM Tag 0018, 9259](https://dicomlookup.com/dicomtags/(0018,9259)) `ASL Crusher Flag`.\n",
"rule": "rules.sidecars.asl.MRIASLCommonMetadataFields"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingOrientation",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Orientation of the labeling plane (`(P)CASL`) or slab (`PASL`).\nThe direction cosines of a normal vector perpendicular to the ASL labeling\nslab or plane with respect to the patient.\nCorresponds to [DICOM Tag 0018, 9255](https://dicomlookup.com/dicomtags/(0018,9255)) `ASL Slab Orientation`.\n",
"rule": "rules.sidecars.asl.MRIASLCommonMetadataFields"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingDistance",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Distance from the center of the imaging slab to the center of the labeling\nplane (`(P)CASL`) or the leading edge of the labeling slab (`PASL`),\nin millimeters.\nIf the labeling is performed inferior to the isocenter,\nthis number should be negative.\nBased on DICOM macro C.8.13.5.14.\n",
"rule": "rules.sidecars.asl.MRIASLCommonMetadataFields"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingLocationDescription",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Description of the location of the labeling plane (`\"CASL\"` or `\"PCASL\"`) or\nthe labeling slab (`\"PASL\"`) that cannot be captured by fields\n`LabelingOrientation` or `LabelingDistance`.\nMay include a link to a deidentified screenshot of the planning of the\nlabeling slab/plane with respect to the imaging slab or slices\n`*_asllabeling.*`.\nBased on DICOM macro C.8.13.5.14.\n",
"rule": "rules.sidecars.asl.MRIASLCommonMetadataFields"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseAverageGradient",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The average labeling gradient, in milliteslas per meter.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseMaximumGradient",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The maximum amplitude of the gradient switched on during the application of\nthe labeling RF pulse(s), in milliteslas per meter.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseAverageB1",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The average B1-field strength of the RF labeling pulses, in microteslas.\nAs an alternative, `\"LabelingPulseFlipAngle\"` can be provided.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseDuration",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Duration of the individual labeling pulses, in milliseconds.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseFlipAngle",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The flip angle of a single labeling pulse, in degrees,\nwhich can be given as an alternative to `\"LabelingPulseAverageB1\"`.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "LabelingPulseInterval",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: Delay between the peaks of the individual labeling pulses, in milliseconds.\n",
"rule": "rules.sidecars.asl.MRIASLCaslPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PCASLType",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The type of gradient pulses used in the `control` condition.\n",
"rule": "rules.sidecars.asl.MRIASLPcaslSpecific"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "EVENTS_TSV_MISSING",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart1_bold.nii.gz",
"rule": "rules.checks.events.EventsMissing"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "CogAtlasID",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart1_bold.nii.gz",
"issueMessage": "Field description: [URI](SPEC_ROOT/common-principles.md#uniform-resource-indicator)\nof the corresponding [Cognitive Atlas](https://www.cognitiveatlas.org/)\nTask term.\n",
"rule": "rules.sidecars.func.MRIFuncTaskInformation"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "CogPOID",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart1_bold.nii.gz",
"issueMessage": "Field description: [URI](SPEC_ROOT/common-principles.md#uniform-resource-indicator)\nof the corresponding [CogPO](http://www.cogpo.org/) term.\n",
"rule": "rules.sidecars.func.MRIFuncTaskInformation"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart1_bold.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart1_bold.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
},
{
"code": "EVENTS_TSV_MISSING",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart2_bold.nii.gz",
"rule": "rules.checks.events.EventsMissing"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "CogAtlasID",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart2_bold.nii.gz",
"issueMessage": "Field description: [URI](SPEC_ROOT/common-principles.md#uniform-resource-indicator)\nof the corresponding [Cognitive Atlas](https://www.cognitiveatlas.org/)\nTask term.\n",
"rule": "rules.sidecars.func.MRIFuncTaskInformation"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "CogPOID",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart2_bold.nii.gz",
"issueMessage": "Field description: [URI](SPEC_ROOT/common-principles.md#uniform-resource-indicator)\nof the corresponding [CogPO](http://www.cogpo.org/) term.\n",
"rule": "rules.sidecars.func.MRIFuncTaskInformation"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PulseSequenceType",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart2_bold.nii.gz",
"issueMessage": "Field description: A general description of the pulse sequence used for the scan.\n",
"rule": "rules.sidecars.mri.MRISequenceSpecifics"
},
{
"code": "SIDECAR_KEY_RECOMMENDED",
"subCode": "PartialFourierDirection",
"severity": "warning",
"location": "/sub-4028/ses-01/func/sub-4028_ses-01_task-SEMTpart2_bold.nii.gz",
"issueMessage": "Field description: The direction where only partial Fourier information was collected.\nCorresponds to [DICOM Tag 0018, 9036](https://dicomlookup.com/dicomtags/(0018,9036)) `Partial Fourier Direction`.\n",
"rule": "rules.sidecars.mri.MRIPartialFourier"
}
],
"codeMessages": {}
},
"summary": {
"sessions": [
"01"
],
"subjects": [
"4028"
],
"subjectMetadata": [],
"tasks": [
"Hypercapnia",
"SEMTpart2",
"SEMTpart1"
],
"modalities": [
"MRI"
],
"secondaryModalities": [
"MRI_Structural",
"MRI_Functional",
"MRI_Perfusion"
],
"totalFiles": 22,
"size": 94580723,
"dataProcessed": false,
"pet": {},
"dataTypes": [
"anat",
"perf",
"func"
],
"schemaVersion": "1.1.3"
}
}
Relevant log outputs (up to 20 lines):
stderr:
slurmstepd: error: Unable to create TMPDIR [/tmp/user/50084]: Permission denied
slurmstepd: error: Setting TMPDIR to /tmp
INFO: Environment variable SINGULARITY_BINDPATH is set, but APPTAINER_BINDPATH is preferred
Process Process-2:
Traceback (most recent call last):
File “/opt/conda/envs/aslprep/lib/python3.11/multiprocessing/process.py”, line 314, in _bootstrap
self.run()
File “/opt/conda/envs/aslprep/lib/python3.11/multiprocessing/process.py”, line 108, in run
self._target(*self._args, **self._kwargs)
File “/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/cli/workflow.py”, line 101, in build_workflow
retval[‘workflow’] = init_aslprep_wf()
^^^^^^^^^^^^^^^^^
File “/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/base.py”, line 69, in init_aslprep_wf
single_subject_wf = init_single_subject_wf(subject_id)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File “/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/base.py”, line 798, in init_single_subject_wf
asl_wf = init_asl_wf(
^^^^^^^^^^^^
File “/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/workflows/asl/base.py”, line 211, in init_asl_wf
run_data = collect_run_data(layout, asl_file)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File “/opt/conda/envs/aslprep/lib/python3.11/site-packages/aslprep/utils/bids.py”, line 106, in collect_run_data
raise FileNotFoundError(f’M0 file for {asl_file} not found.')
FileNotFoundError: M0 file for /bids/sub-4028/ses-01/perf/sub-4028_ses-01_asl.nii.gz not found.
stdout
Using workdir: /data/raulab/asl2/aslwd
260108-01:19:44,209 nipype.workflow IMPORTANT:
Running ASLPrep version 25.1.0
License NOTICE ##################################################
ASLPrep 25.1.0
Copyright 2023 The PennLINC Team and the NiPreps Developers.
This product is primarily developed by the PennLINC team,
but it is also a part of the NiPreps community.
This product includes software developed by
the NiPreps Community (https://nipreps.org/).
Portions of this software were developed at the Department of
Psychology at Stanford University, Stanford, CA, US.
This software is also distributed as a Docker container image.
The bootstrapping file for the image ("Dockerfile") is licensed
under the MIT License.
This software may be distributed through an add-on package called
"Docker Wrapper" that is under the BSD 3-clause License.
#################################################################
260108-01:19:48,595 nipype.workflow IMPORTANT:
Building ASLPrep's workflow:
* BIDS dataset path: /bids.
* Participant list: ['4028'].
* Run identifier: 20260108-011903_94843686-6d24-422c-9061-9733e66f0f2d.
* Output spaces: T1w MNI152NLin2009aSym:res-native.
* Pre-run FreeSurfer's SUBJECTS_DIR: /out/sourcedata/freesurfer.
260108-01:19:49,955 nipype.workflow INFO:
ANAT Stage 1: Adding template workflow
260108-01:19:50,909 nipype.workflow INFO:
ANAT Stage 2: Preparing brain extraction workflow
260108-01:19:51,66 nipype.workflow INFO:
ANAT Stage 3: Preparing segmentation workflow
260108-01:19:51,76 nipype.workflow INFO:
ANAT Stage 4: Preparing normalization workflow for ['MNI152NLin2009aSym', 'MNI152NLin2009cAsym']
260108-01:19:51,102 nipype.workflow INFO:
ANAT Stage 5: Preparing surface reconstruction workflow
260108-01:19:51,145 nipype.workflow INFO:
ANAT Stage 6: Preparing mask refinement workflow
260108-01:19:51,151 nipype.workflow INFO:
ANAT Stage 7: Creating T2w template
260108-01:19:51,167 nipype.workflow INFO:
ANAT Stage 8: Creating GIFTI surfaces for ['white', 'pial', 'midthickness', 'sphere_reg', 'sphere']
260108-01:19:51,210 nipype.workflow INFO:
ANAT Stage 8: Creating GIFTI metrics for ['thickness', 'sulc']
260108-01:19:51,227 nipype.workflow INFO:
ANAT Stage 8a: Creating cortical ribbon mask
260108-01:19:51,234 nipype.workflow INFO:
ANAT Stage 9: Creating fsLR registration sphere
260108-01:19:51,244 nipype.workflow INFO:
ANAT Stage 10: MSM-Sulc disabled
260108-01:19:51,244 nipype.workflow INFO:
ANAT Stage 11: Creating cortical surface mask
Screenshots / relevant information: