Summary of what happened:
Dear all,
I am trying to analyze data from the MB EPI PCASL Sequence (from Minnesota University) using oxford_asl.
I found in an old conversation in jiscmail (“Oxford ASL and MB-pCASL”) some flags, which I used, but still the results are not satisfying…
We first set up the sequence using the same parameters as used in the HCP protocol. The images from the scanner look overall good.
I then extracted the first 86 volumes as input image and the last 2 as calibration one.
Command used (and if a helper script was used, a link to the helper script or the command generated):
Here the call for oxford_asl with the flags I used:
oxford_asl \
-i "input_file.nii.gz" \
-o "output" \
--casl \
--iaf tc \
--ibf tis \
--tis 1.7,2.3,2.8,3.3,3.8 \ #i.e. 0.2 + 1.5 (bolus duration), 0.8+1.5, 1.3+1.5, ...
--rpts 6,6,6,10,15 \ #same as I found on my .ini file
--bolus 1.5 \
--slicedt 0.0045 \
--fslanat "struc.anat" \
--pvcorr \
-c "calibration_image.nii.gz" \
--tr 3.6 \ #<-- in the past conversation it was mentioned a tr=8, but I checked my calibration image and the tr is 3.6
--mc \
Version:
Environment (Docker, Singularity, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
Relevant log outputs (up to 20 lines):
Screenshots / relevant information:
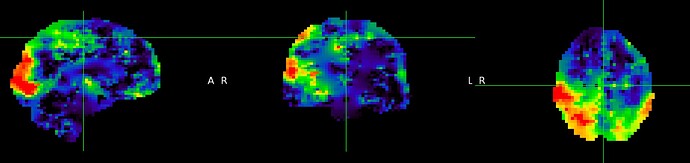
Attached a screenshot of the perfusion_calib image after pvcorr (I’m sorry, I am new here and I still don’t know how to upload .nii files here). What I see is that the perfusion map doesn’t reflect the GM and WM perfusion. Moreover, I obtained a perfusion_calib in gm is 4.007116 (too small).
Does anybody know what could be the problem? My guess is that some flags are wrong.
Your help would be highly appreciated.
Lorenzo Nigris