Sorry if this isn’t the right place for this sort of question.
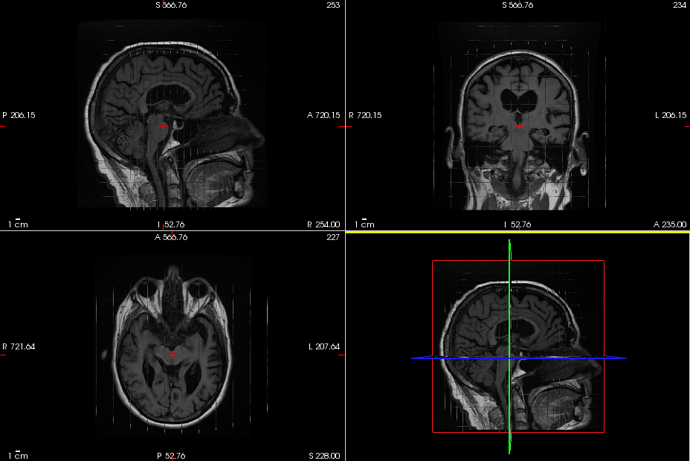
Wondering if anyone here is familiar with the clinical practice of taking anatomical MRIs in the three directions separately. This results in very thick slices with high spatial resolution in the two orthogonal directions…
Our clinical scans have three of these, one for each direction, and I was curious if it’s possible to take these three scans and “merge” them into one image with higher resolution in all directions.
I’m not familiar with this practice, but assuming that these images are already aligned, you could probably do something like:
import numpy as np
from nilearn.image import reorder_img, resample_img, math_img
# Load and reorient if needed to ensure consistent RAS orientation
x = reorder_img('thick_x.nii.gz')
y = reorder_img('thick_y.nii.gz')
z = reorder_img('thick_z.nii.gz')
# Affines should be the same over the two of three columns
tgt_affine = np.median((x.affine, y.affine, z.affine), axis=0)
# Shapes should be the same over two of three dimensions
tgt_shape = tuple(np.median((x.shape, y.shape, z.shape), axis=0).astype(int))
# You may add interpolation='nearest' to avoid interpolation, but this will
# increase blockiness
res_x = resample_img(x, target_affine=tgt_affine, target_shape=tgt_shape)
res_y = resample_img(y, target_affine=tgt_affine, target_shape=tgt_shape)
res_z = resample_img(z, target_affine=tgt_affine, target_shape=tgt_shape)
# Alternately you can try median
new_img = math_img('np.mean((x, y, z), axis=0)', x=res_x, y=res_y, z=res_z)
I tested this by taking an existing T1 image (voxels: 1mm x 1.3mm x 1.3mm), and I was able to get a reasonably good reconstruction, but it definitely does not restore the full resolution at each point. I don’t know how to estimate the effective resolution of this image; it’s going to be better near the middle of your thick slices, where you have high-quality source information, and worse as you get to the grid corners farthest away from any mid-slice plane, where we have to interpolate in all three directions.
You may be able to reduce distortion by using nearest-neighbor interpolation (rather than the default continuous) and the median to combine your results, but it will result in blockiness. It’s a question of whether you want information fidelity or something nice to look at.
For posterity, Eli and I work together and had some time to kill.
We had a few scans with this type of data and tested it. For our images, the command
x = reorder_img(‘thick_x.nii.gz’)
gave an error involving rotation. We weren’t super familiar with nilearn and couldn’t resolve it.
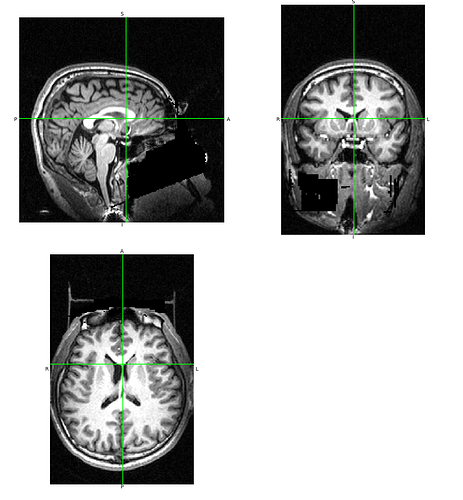
We did a similar algorithm using MATLAB to some success, and the images one gets out are illustrative of how little data one obtains in a scan like this.
Essentially the idea was to think of the image space as a grid–each scan gives measurements of one slice of that grid with great accuracy, but then skips many in a given direction. By extending each image out with zeros and then averaging the non-zero measurement for each voxel, one could in theory obtain a measurement of many more nodes in the grid.
Here’s an image with no interpolation:
Essentially you have the “nice slices” in each direction, and a whole lot of black space in between. The images dimensions were permutations of 512x512x22, so the amount of up sampling necessary means there’s actually way less information in these scans than one might think a priori.
There’s some subtleties of field of view in one dimension that I couldn’t figure out in the free afternoon I have, so it would be a little bit better if we could fix that.
1 Like
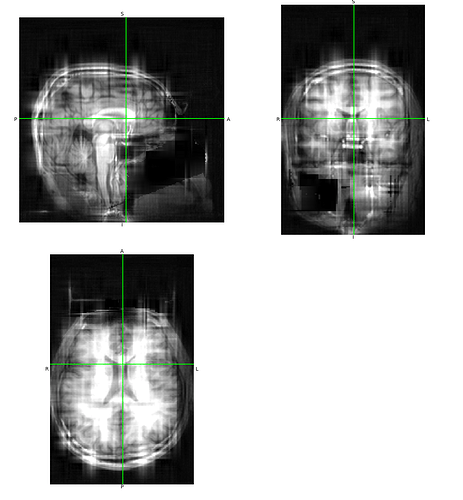
Yeah, I was trying this with a 5:1 down-sampling. 23:1 will give you much more distortion away from the mid-slice planes.
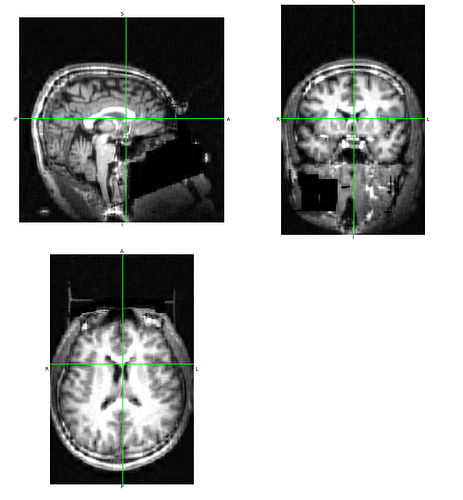
For reference, here is the original:
And “reconstructed”:
Trying it again at a 20:1 ratio:
So yeah, you can get something to look at, at that ratio, but the low source resolution results in a ton of artifacts.