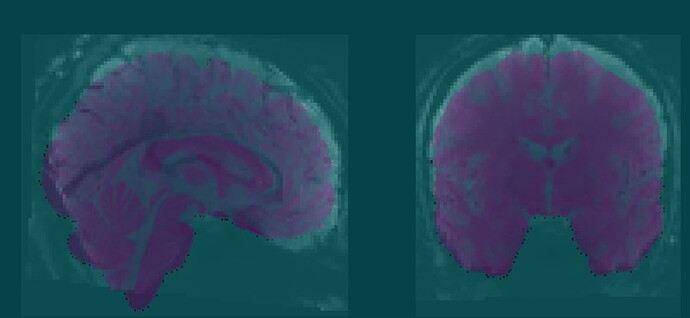
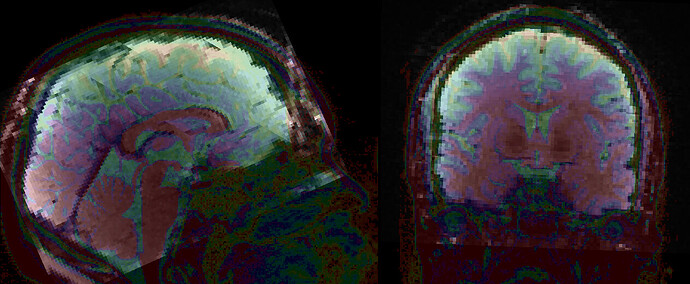
Summary of what happened:
Hi fMRIPrep experts,
While performing statistical analysis, I noticed NaN values in the GM mask for some subjects. Upon reviewing the fMRIPrep output (desc-preproc_bold.nii.gz) , I discovered that this was due to misalignment in the fMRI data in MNI space. Specifically, one of the two BOLD runs was not properly aligned, whereas the other run showed good alignment, suggesting that the normalization between native space and MNI space was good.
The mismatch became more apparent when switching between the two runs. Then I suspected a severe mismatch between this run and the T1w in the raw data, which could have caused the within-subject alignment to fail. However, the mismatch was minor and appeared typical for pre-alignment.
This issue wasn’t limited to just one or two subjects—it affected about 10% of my dataset. I have uploaded the BIDS and some fMRIPrep output for one subject for reference.
https://www.dropbox.com/scl/fo/1tvw5iiovzvxchm3s8upd/AMA9MtPzekcWcbqKHe6p5W8?rlkey=al8jflsyos2wpce41bjh7m4fl&st=b3pqytm2&dl=0
my fMRIPrep command
export SINGULARITY_BIND=$outpath
singularity run --cleanenv /home/toolbox/fMRIPrep/nipreps_fmriprep_24.0.1-2024-07-16-d329b3eeee9f.simg \
$bidspath $outpath \
participant --participant-label ${subID[$SLURM_ARRAY_TASK_ID]} \
--work-dir $outpath/workingD/ \
--fs-license-file /home/toolbox/freesurfer_license/license.txt \
--write-graph \
--output-spaces T1w MNI152NLin2009cAsym fsaverage fsaverage5 fsaverage6 \
--cifti-output 91k --project-goodvoxels \
--fs-subjects-dir $fspath \
--fs-no-resume
Version:
fmriprep_24.0.1-2024-07-16
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity on google cloud
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE