Summary of what happened:
Hello,
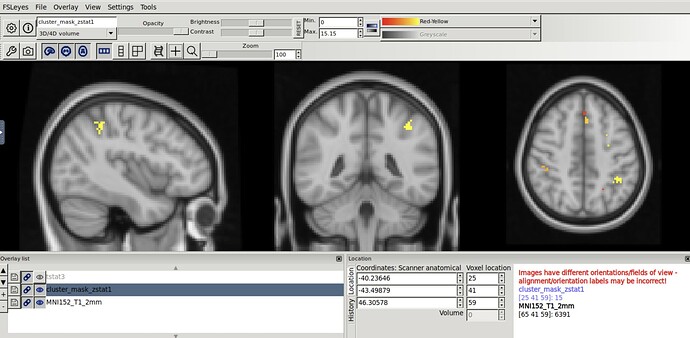
I used fMRIprep to preprocess some functional task data. Then, I ran stats using FSL’s feat (following this Mumford Brain Stats — FEAT registration workaround to deal with registration stuff). When I ran fMRI prep, I used the output space MNI152NLin6Asym, which is believe is the standard space for FSL. However, when I open my contrast results on top of the FSL standard brain, the voxel numbers do not match up. It seems like perhaps there was a left-right flip at some point.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fmriprep $ROOT $OUT participant --participant-label $SUB --mem 128000 --work-dir $WORK --output-space MNI152NLin6Asym:res-2 fsLR:den-32k --fs-license-file $FS_LICENSE --fd-spike-threshold 1 --stop-on-first-crash
Version:
23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE