ssinha
February 13, 2025, 11:03pm
1
Hello,
I wanted to reach out regarding two questions I have about generating correlation matrices via XCP-D.
In my output folder (~sub-ECDMT40000/ses-1/func), it looks like many of the file types mentioned in this link from the documentation are missing. After checking, it looks the following file types were not generated:
NIfTI: <source_entities>_space-<label>_seg-<label>_stat-pearsoncorrelation_relmat.tsvCIFTI: <source_entities>_space-fsLR_seg-<label>_den-91k_stat-coverage_bold.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-mean_timeseries.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_relmat.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_desc-<INT>volumes_relmat.tsv
Should I include any additional arguments or flags in my command to generate these above file types?
My second question is about the value of the CORRELATION_LENGTHS that goes with the --create-matrices argument. Is there any upper or lower limit that I should keep in mind for this value?
Thanks,
singularity run \
-e -B /scratch1/sinha/ECDMT40000/derivatives \
-B /scratch1/sinha/xcp_output_02132025_ln10 \
/data/nil-external/ccp/XCP-D/xcp_d_dec2024.simg \
/scratch1/sinha/ECDMT40000/derivatives \
/scratch1/sinha/xcp_output_02132025_ln10 \
participant \
--smoothing 2 \
--dummy-scans 6 \
--random-seed 0 \
--bpf-order 2 \
--despike \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
-p 36P \
--motion-filter-type notch \
--band-stop-min 15 \
--band-stop-max 25 \
--motion-filter-order 4 \
--head-radius auto \
--file-format cifti \
--warp-surfaces-native2std \
--mode abcd \
--abcc-qc y \
--create-matrices 10 \
--atlases 4S456Parcels \
--fd-thresh 0.3
XCP-D v0.10.1
XCP-D was run as a singularity docker image (.simg file) in a computing cluster.
PASTE VALIDATOR OUTPUT HERE
PASTE LOG OUTPUT HERE
Steven
February 13, 2025, 11:14pm
2
Hi @ssinha ,
Did the job proceed without error? Does the behavior persist in most recent verison (0.10.5)? Can you provide a tree directory structure of a subject’s xcp_d outputs? Is this true for all subjects? Anything helpful in the log file?
Best,
ssinha
February 14, 2025, 6:16pm
3
Thanks, Steven. Here are my responses:
The job was successfully completed without any error messages.
Our lab has not yet used the most recent version. The one I am working with is XCP-D v0.10.1.
I found that the tree command is not available in the cluster. I found a workaround though and here is the full structure:
.
|-logs
| |-CITATION.md
| |-CITATION.bib
| |-CITATION.tex
| |-CITATION.html
|-atlases
| |-dataset_description.json
| |-atlas-4S456Parcels
| | |-atlas-4S456Parcels_space-fsLR_den-91k_dseg.dlabel.nii
| | |-atlas-4S456Parcels_space-fsLR_den-91k_dseg.json
| | |-atlas-4S456Parcels_dseg.tsv
|-sub-ECDMT40000_ses-1_executive_summary.html
|-dataset_description.json
|-sub-ECDMT40000.html
|-desc-linc_qc.json
|-sub-ECDMT40000
| |-figures
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT1wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnat_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialSuperiorFrontal_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlasBrainSwipes_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-boldref_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialSuperiorFrontal_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT1w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-postprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-rehoSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT2wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-mean_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalPosteriorParietalLingual_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT2w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-preprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T1wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalCorpusCallosum_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-postprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-alffSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-mosaic_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalOrbitoFrontal_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalOrbitoFrontal_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-alffSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-qualitycontrol_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT2w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-preprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialBasalGangliaPutamen_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnatBrainSwipes_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalCaudateAmygdala_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-bbregister_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlas_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaTemporalHippocampalSulcus_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT2wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T2wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-rehoSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaTemporalHippocampalSulcus_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalCaudateAmygdala_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-coverageParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T2wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-postprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T2wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-alffParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlasBrainSwipes_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-censoring_motion.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-boldref_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-mosaic_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnatBrainSwipes_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-about_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-rehoParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-summary_bold.html
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaFrontoTemporal_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-preprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialBasalGangliaPutamen_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T1wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalPosteriorParietalLingual_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-qualitycontrol_bold.html
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlas_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-postprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-mean_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-rehoParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-bbregister_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaFrontoTemporal_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-preprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialInferiorTemporalCerebellum_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialInferiorTemporalCerebellum_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T2wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T1wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T1wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnat_T1w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT1wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT1w_bold.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalCorpusCallosum_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-coverageParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-alffParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-censoring_motion.svg
| |-ses-1
| | |-func
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_desc-abcc_qc.hdf5
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-linc_qc.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_desc-abcc_qc.hdf5
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_relmat.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-linc_qc.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_outliers.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-reho_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_motion.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.pscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-reho_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_outliers.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_motion.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.ptseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-reho_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoised_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoised_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoised_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_motion.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-reho_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_outliers.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_relmat.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.pscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_motion.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoised_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_outliers.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.ptseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.json
| | |-anat
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_curv.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_thickness.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_inflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_sulc.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_midthickness.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-curv_morph.tsv
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_inflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_white.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-preproc_T2w.nii.gz
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_midthickness.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_vinflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-sulc_morph.tsv
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_white.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-thickness_morph.tsv
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_vinflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_pial.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-preproc_T1w.nii.gz
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_pial.surf.gii
| |-log
| | |-20250213-145728_e9abf877-d605-4dec-a129-e878b0504590
| | | |-xcp_d.toml
I have ran separate trials with 4 different subjects as of now, and they all have the same file-types missing.
There are some flags set to false but other than that, I did not notice anything significant in the log file.
tsalo
February 17, 2025, 2:23pm
4
If you could share the stdout and stderr files from your processing job, I think that would help. Also, can you sort the items in your tree output? It’s really hard to make sense of it, since it looks like the files are in a random order.
I noticed that you have --create-matrices 10 in your call. This will produce a correlation matrix from a randomly-selected 10 seconds of data. Is that what you want?
ssinha
February 17, 2025, 9:53pm
5
It looks like the stdout and stderr files are not present in the /proc folder. I will try to re-run another job today and see if I can find it. I do have the -xcp_d.toml file and also saved the output messages in a .txt file if you need it.
Here is the sorted version of the directory structure:
.
|-atlases
| |-atlas-4S456Parcels
| | |-atlas-4S456Parcels_dseg.tsv
| | |-atlas-4S456Parcels_space-fsLR_den-91k_dseg.dlabel.nii
| | |-atlas-4S456Parcels_space-fsLR_den-91k_dseg.json
| |-dataset_description.json
|-dataset_description.json
|-desc-linc_qc.json
|-logs
| |-CITATION.bib
| |-CITATION.html
| |-CITATION.md
| |-CITATION.tex
|-sub-ECDMT40000
| |-figures
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlasBrainSwipes_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlasBrainSwipes_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlas_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AnatOnAtlas_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnatBrainSwipes_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnatBrainSwipes_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnat_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AtlasOnAnat_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialBasalGangliaPutamen_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialBasalGangliaPutamen_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialInferiorTemporalCerebellum_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialInferiorTemporalCerebellum_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialSuperiorFrontal_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-AxialSuperiorFrontal_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalCaudateAmygdala_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalCaudateAmygdala_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalOrbitoFrontal_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalOrbitoFrontal_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalPosteriorParietalLingual_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-CoronalPosteriorParietalLingual_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-mosaic_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-mosaic_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalCorpusCallosum_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalCorpusCallosum_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaFrontoTemporal_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaFrontoTemporal_T2w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaTemporalHippocampalSulcus_T1w.png
| | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-SagittalInsulaTemporalHippocampalSulcus_T2w.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-about_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-alffParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-alffSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-censoring_motion.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-coverageParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-postprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-postprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-preprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-preprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-qualitycontrol_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-rehoParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-rehoSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_desc-summary_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-bbregister_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-boldref_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-mean_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T1wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T1wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T2wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-T2wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT1w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT1wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT2w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-MNI152NLin6Asym_desc-TaskOnT2wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-alffParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-alffSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-censoring_motion.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-coverageParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-postprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-postprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-preprocESQC_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-preprocessing_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-qualitycontrol_bold.html
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-rehoParcellatedSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_desc-rehoSurfaceSubject_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-bbregister_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-boldref_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-mean_bold.svg
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T1wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T1wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T2wOnTask_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-T2wOnTaskBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT1w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT1wBrainSwipes_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT2w_bold.png
| | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-MNI152NLin6Asym_desc-TaskOnT2wBrainSwipes_bold.png
|-sub-ECDMT40000.html
| |-log
| | |-20250213-145728_e9abf877-d605-4dec-a129-e878b0504590
| | | |-xcp_d.toml
| |-ses-1
| | |-anat
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_inflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_midthickness.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_desc-hcp_vinflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_pial.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-L_space-fsLR_den-32k_white.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_inflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_midthickness.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_desc-hcp_vinflated.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_pial.surf.gii
| | | |-sub-ECDMT40000_ses-1_hemi-R_space-fsLR_den-32k_white.surf.gii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_curv.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_sulc.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_den-91k_thickness.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-curv_morph.tsv
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-sulc_morph.tsv
| | | |-sub-ECDMT40000_ses-1_space-fsLR_seg-4S456Parcels_stat-mean_desc-thickness_morph.tsv
| | | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-preproc_T1w.nii.gz
| | | |-sub-ECDMT40000_ses-1_space-MNI152NLin6Asym_desc-preproc_T2w.nii.gz
|-sub-ECDMT40000_ses-1_executive_summary.html
| | |-func
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_desc-abcc_qc.hdf5
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_design.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_motion.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_motion.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_outliers.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_outliers.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoised_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoised_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_desc-linc_qc.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-reho_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_stat-reho_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.pscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.ptseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_relmat.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_desc-abcc_qc.hdf5
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_design.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_motion.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_motion.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_outliers.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_outliers.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoised_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoised_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.dtseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-denoisedSmoothed_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_desc-linc_qc.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-alff_desc-smooth_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-reho_boldmap.dscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_den-91k_stat-reho_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-coverage_boldmap.pscalar.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-mean_timeseries.ptseries.nii
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-alff_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_relmat.tsv
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.json
| | | |-sub-ECDMT40000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-reho_bold.tsv
Thanks for letting me know about the correlation length flag in my --create-matrices argument. I was not sure about the usage of the correlation length flag and chose a value of 10 to start with. Ideally, I would like to generate the matrices over the entire data.
tsalo
February 17, 2025, 9:57pm
6
In the new list of files, it looks like you have the outputs you wanted.
In that case, you want --create-matrices all.
ssinha
February 17, 2025, 10:03pm
7
These file-types are missing from the ~sub-ECDMT40000/ses-1/func folder:
<source_entities>_space-<label>_seg-<label>_stat-pearsoncorrelation_relmat.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-coverage_bold.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-mean_timeseries.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_relmat.tsv<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_desc-<INT>volumes_relmat.tsv
Should I use any additional parameters to generate these above file-types?
tsalo
February 18, 2025, 3:28pm
8
You are getting some of those files:
sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_relmat.tsv
sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-coverage_bold.tsv
sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-mean_timeseries.tsv
You won’t get correlation matrices without the desc-<INT>volumes entity because you didn’t use --create-matrices all, but other than that your expected outputs mostly match what you get. Based on your settings, the only one you seem to be missing is
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii
which would be named
sub-ECDMT40000_ses-1_task-MOVIEdm_run-1_space-fsLR_den-91k_seg-4S456Parcels_stat-pearsoncorrelation_desc-12volumes_boldmap.pconn.nii```
I’m not sure why that file is missing, but if you could rerun your job with it set up to write out the output and error log files, and then share them, that would make it easier to figure out.
ssinha
February 18, 2025, 8:08pm
9
Thanks for letting me know.
I reran the job with the --create-matrices all argument. Here’s the full command I used:
singularity run \
-e -B /scratch1/sinha/ECDMT40010/derivatives \
-B /scratch1/sinha/xcp_output_02172025 \
/data/nil-external/ccp/XCP-D/xcp_d_dec2024.simg \
/scratch1/sinha/ECDMT40010/derivatives \
/scratch1/sinha/xcp_output_02172025 \
participant \
--smoothing 2 \
--dummy-scans 6 \
--random-seed 0 \
--bpf-order 2 \
--despike \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
-p 36P \
--motion-filter-type notch \
--band-stop-min 15 \
--band-stop-max 25 \
--motion-filter-order 4 \
--head-radius auto \
--file-format cifti \
--warp-surfaces-native2std \
--mode abcd \
--abcc-qc y \
--create-matrices all \
--atlases 4S456Parcels \
--fd-thresh 0.3
I checked the output and it looks like the pconn.nii filetypes were generated this time. However, I still have the following filetypes missing.
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-coverage_bold.tsv
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-mean_timeseries.tsv
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_relmat.tsv
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_desc-<INT>volumes_relmat.tsv
Is there anything that could be missing from my command or preempting the creation of these filetypes? Regarding setting up the output and error log files to be written, is there any specific way through which this needs to be implemented?
tsalo
February 18, 2025, 8:16pm
10
I believe you have those, they just don’t have den-91k in the filename.
Also, you won’t get stat-pearsoncorrelation_desc-<INT>volumes_relmat.tsv since you used --create-matrices all without any exact-time values.
ssinha
February 21, 2025, 8:48pm
11
Thanks for your prior assistance. I have two quick follow-up questions on this topic:
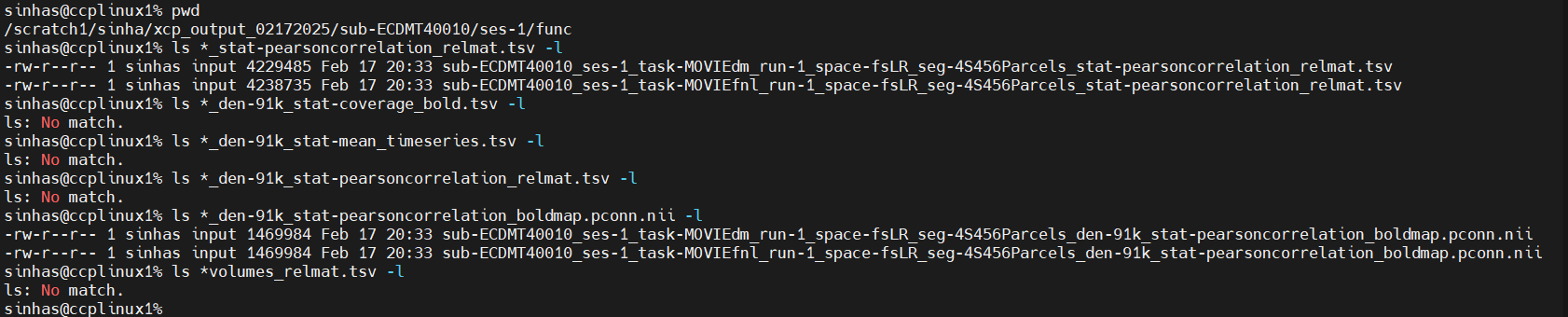
My first question is about the following three file-types. I have run many trials as of now with different subjects, but these are still missing. This link in the documentation says I am supposed to have these files. However, I do have the “non-91k” versions of these files. Should I ignore these file-types?
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-coverage_bold.tsv
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-mean_timeseries.tsv
<source_entities>_space-fsLR_seg-<label>_den-91k_stat-pearsoncorrelation_relmat.tsv
My second question is about the *pconn.nii files such as these two that I recently generated with XCP-D:
~/sub-ECDMT30000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii
~/sub-ECDMT30000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii
I was able to visualize these in connectome workbench, but I was wondering about the values they represent. For instance, is there only one “frame” in the fall and does this reflect the average connectivity for each parcel with all other parcels? Or are their many frames, and each one reflect the connectivity of a different parcel with all other parcels?
ssinha
March 10, 2025, 8:21pm
12
I wanted to follow up on this topic regarding the two questions I had.
My first question was about the remaining three missing file-types (pasted in the earlier response). I was wondering whether we should ignore these or if there are any changes that I should incorporate in my command to generate these.
My second question was about the difference between the two variants (task-MOVIEdm & task-MOVIEfnl ) of the file-types that are generated as output. We are wondering if there is only one frame in the fall and if it reflects the average connectivity for each parcel with all other parcels, or if there are many frames, such that each one reflects the connectivity of a different parcel with all other parcels.
sub-ECDMT30000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.json
sub-ECDMT30000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii
sub-ECDMT30000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_relmat.json
sub-ECDMT30000_ses-1_task-MOVIEdm_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_relmat.tsv
sub-ECDMT30000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.json
sub-ECDMT30000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_den-91k_stat-pearsoncorrelation_boldmap.pconn.nii
sub-ECDMT30000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_relmat.json
sub-ECDMT30000_ses-1_task-MOVIEfnl_run-1_space-fsLR_seg-4S456Parcels_stat-pearsoncorrelation_relmat.tsv
tsalo
March 10, 2025, 8:28pm
13
The non-91k is the same thing as the 91k version.
I believe each volume in the pconn file reflects a single parcel’s connectivity to each of the other parcels.
Those aren’t variants created by XCP-D. They’re different tasks in your BIDS dataset. I assume they represent different movies presented to the participants.
This one is correct, as far as I know. I don’t personally use the pconn files, but that’s my understanding.
1 Like