We recently ran fMRIPrep on two sets of resting-state open-source fMRI data. The first set is the UCLA (Consortium for Neuropsychiatric Phenomics study, site: https://www.openfmri.org/dataset/ds000030/) and the second set is the COBRE (International Neuroimaging Data-sharing Initiative) dataset (site: http://fcon_1000.projects.nitrc.org/indi/retro/cobre.html).
Upon examining the fMRIPrep preprocessed fMRI data, we found that the fMRI in MNI spaces (both NLin6 and NLin2009c) looked incorrectly normalized compared with data preprocessed with SPM12.
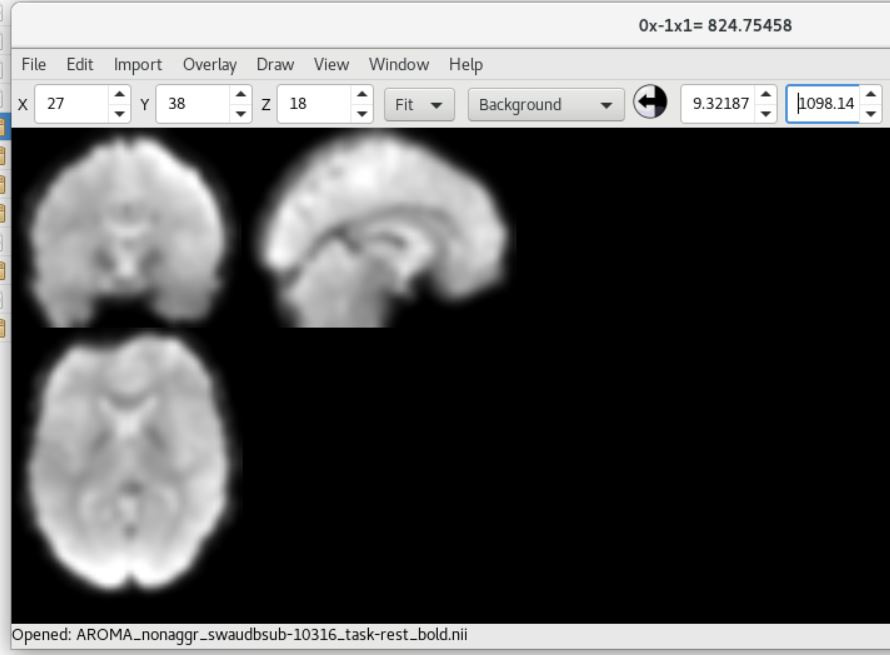
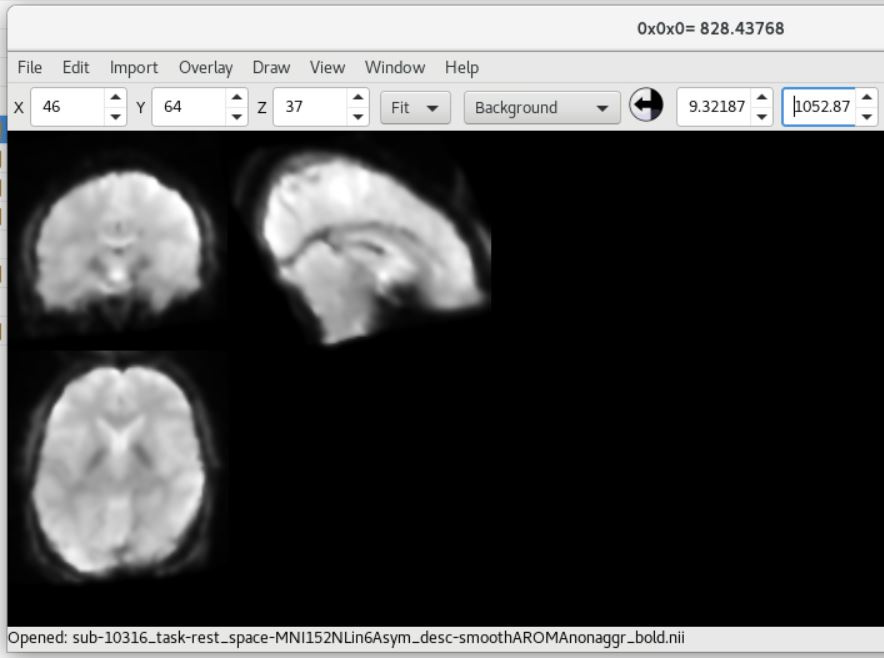
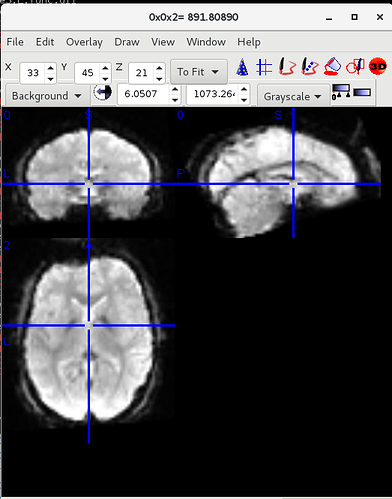
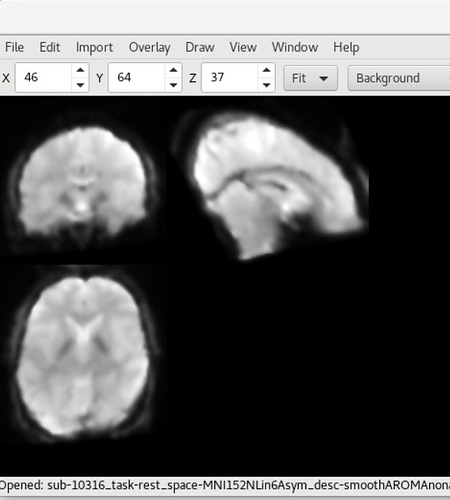
Here is the UCLA data showing SPM12 (top) and fMRIPrep (bottom) outputs (from sub-10316)
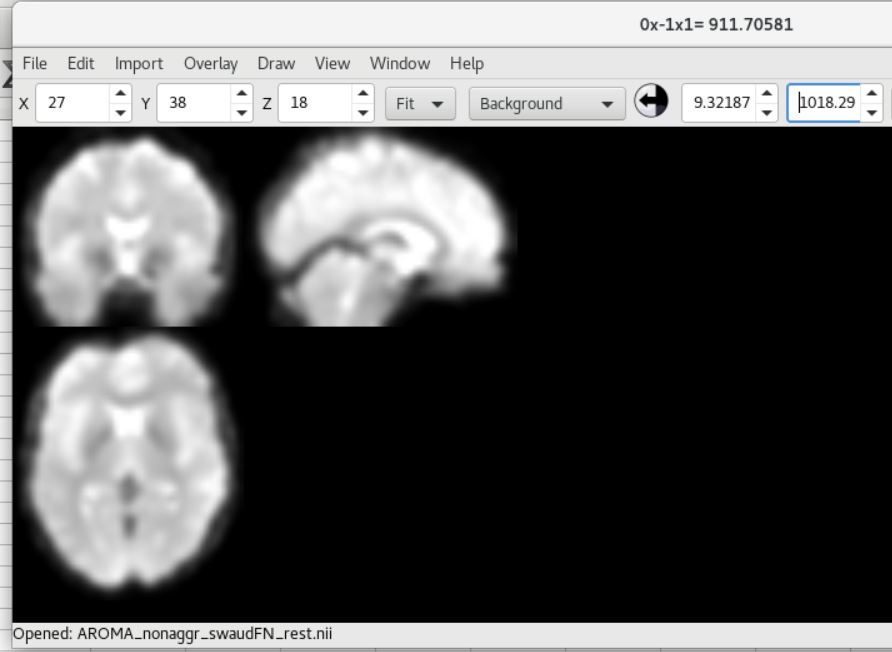
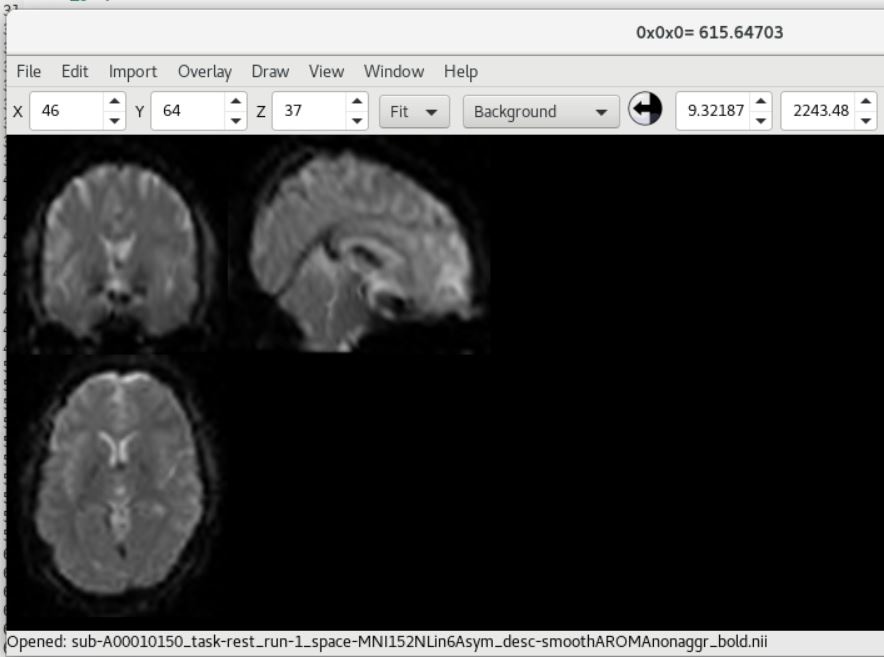
And here is the COBRE data showing SPM12 (top) and fMRIPrep (bottom) outputs (from subject ID 0040024 aka A00010150).
As you can see, there is a strong forward sagittal tilt and coronal skewness in the fMRIPrep preprocessed data that does not appear in the SPM12 preprocessed data. The T1 weighted anatomical data (not shown) did appear to normalize correctly for both datasets with fMRIPrep, but matched poorly with the normalized functional data.
I should note that I have used fMRIPrep successfully on our ‘in-house’ fMRI data at the Olin Neuropsychiatry Research Center (with different sequences/TRs) without these normalization issues.
We used fMRIPrep version 20.1.1 to preprocess both datasets on two servers (different locations) at with separate fMRIPrep installations. In various tests, the normalization problems with fMRIPrep did not appear to depend on settings such as slice timing (on or off), SDC (on or off, with field maps or fieldmap-less).
Does anyone know what might be causing these problems with these two datasets? Is it possible there is information in BIDS JSON sidecar files from these two datasets that would cause these problems?
Thanks for any help you can provide.
Chris