I would strongly suggest using the NIfTI SForm and QForm to infer spatial orientation of the data. Making a hard-coded assumption regarding the data is asking for unintended consequences. While it might take a little more work up front, the long term code maintenance will be much better.
For example, with this image
>fslhd l/fsl/data/standard/MNI152lin_T1_2mm.nii.gz
filename MNI152lin_T1_2mm.nii.gz
size of header 348
data_type UINT8
dim0 3
dim1 91
dim2 109
...
sform_name MNI_152
sform_code 4
sto_xyz:1 -2.000000 0.000000 0.000000 90.000000
sto_xyz:2 0.000000 2.000000 0.000000 -126.000000
sto_xyz:3 0.000000 0.000000 2.000000 -72.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
The Form suggests that the data is stored on disk LAS (e.g. first voxel on disk is on the Right side of the brain, proceeding Left-ward). While the data below is stored RAS.
sform_name MNI_152
sform_code 4
sto_xyz:1 2.000000 0.000000 0.000000 -90.000000
sto_xyz:2 0.000000 2.000000 0.000000 -126.000000
sto_xyz:3 0.000000 0.000000 2.000000 -72.000000
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Left-to-Right
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
Personally, I use the terms “Radiological” and “Neurological” to describe conventions for viewing data on a screen, not storing data to disk. A radiologist assumes they are viewing slices from the patient’s feet, the way we see a participant from the MRI console room while they are getting scanned, hence their left is on the viewer’s right. In contrast, a neurologist assumes they have a ‘birds eye’ view of the head from above. This explains why coronal and axial slices appear flipped between these two views, but surface renderings do not.
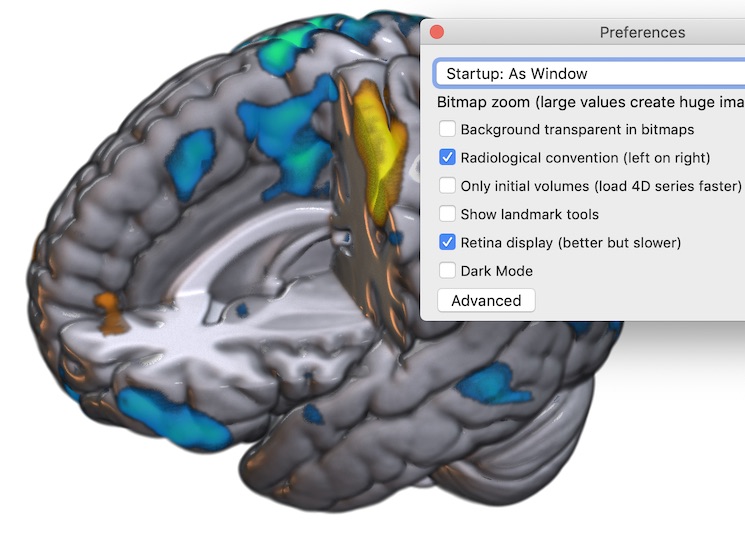
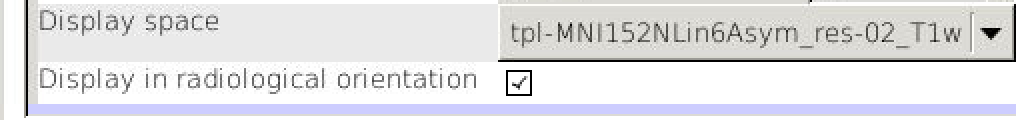
FSLview makes this distinction clear, if you click on the “Gear” (“Spanner”) icon you can choose whether “Display Space” is “World” or disk orientation, but there is also a checkbox to toggle between radiological and neurological convention. Likewise, while MRIcroGL always shows images oriented to world-space, it allows the user to choose between viewing conventions (see below).
tldr; but for completeness: One final word of caution when working with NIfTI files. There are actually two ways of defining space: a spatial matrix (the SForm) and a Quaternion (the QForm). In theory, one could use these two independently: the QForm could be the data as acquired on the scanner while the SForm could encode a linear warp to normalized space (e.g. modeling the influence of rotations, zooms, translations and shears). Unfortunately, VTK based tools gave precedence to the QForm (ANTS, slicer, etc), while most other tools gave precedence to the SForm. Therefore, to switch between tools I would always try to keep these two the same, and think carefully about what to do when they disagree. While there are simple transforms between the SForm and the QForm, you need to be mindful that in theory a spatial matrix is a superset of the quaternion. A quaternion can only store translations, rotations and zooms. The matrix can store the full affine set: translations, rotations, zooms and shears. Therefore, any quaternion can be losslessly represented as a matrix, but the reverse is not true. I suspect this is why VTK adopted the QForm precedence: VTK volumes must be rectangular and therefore cannot abide by shears. Fortunately, while shears are common for CT (gantry tilt), they are rare in MRI. Even a full 12-DoF ‘affine transform’ from FLIRT does not model shears.