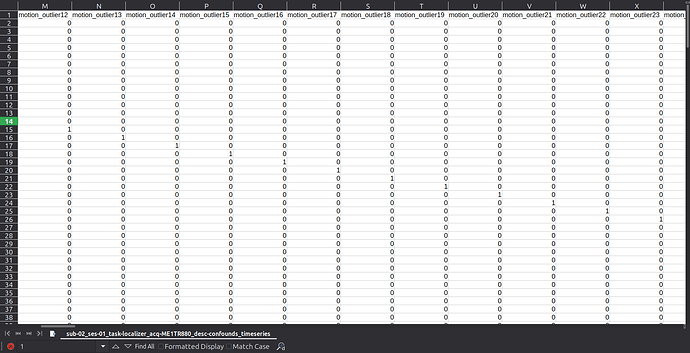
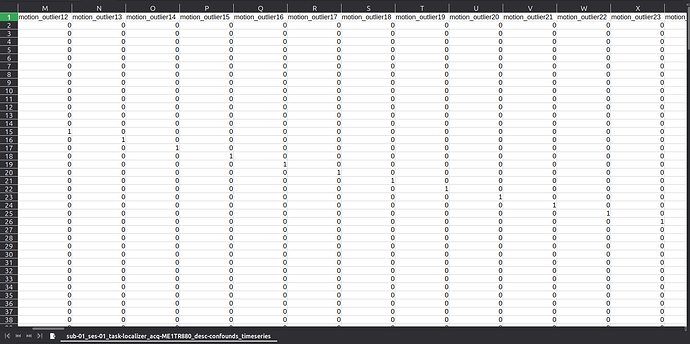
Summary of what happened:
Hello, we’ve noticed an unexpected pattern in the motion_outlier columns of the confounds timeseries output. For each scan, we have an outlier detected even though no apparent movement is visible when inspecting raw data. The issue appears consistently across different functional sequences and subjects.
I would appreciate any insight on what might be causing this. Please find attached the timeseries files with unrelated columns removed to make the issue easier to see. I also attach a screenshot of the same files for a subset of the motion_outlier columns for easier visualization of the problem.
Command used (and if a helper script was used, a link to the helper script or the command generated):
FMRIPREP="my_images/fmriprep-25.2.4.simg"
singularity run --cleanenv \
-B $bids_root_dir:/data \
"$FMRIPREP" \
/data/sourcedata/raw /data/derivatives \
participant \
--participant-label "$subj" \
--nprocs 16 \
--omp-nthreads 16 \
--level full \
--output-spaces MNI152NLin2009cAsym anat \
--me-t2s-fit-method loglin \
--fs-license-file /data/code/license.txt \
--stop-on-first-crash \
--work-dir /data/work \
-vv
Version:
25.2.4
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
The data is BIDS compliant.
Screenshots / relevant information:
sub-02_ses-01_task-localizer_acq-ME1TR880_desc-confounds_timeseries.txt (97.1 KB)
sub-01_ses-01_task-localizer_acq-ME1TR880_desc-confounds_timeseries.txt (105.0 KB)
sub-02_ses-01_task-localizer_acq-DresdenWFat_desc-confounds_timeseries.txt (11.2 KB)
sub-01_ses-01_task-localizer_acq-DresdenWFat_desc-confounds_timeseries.txt (13.0 KB)