Hi-
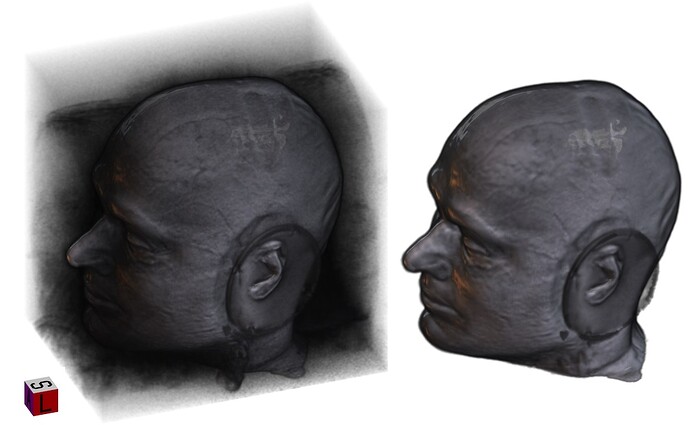
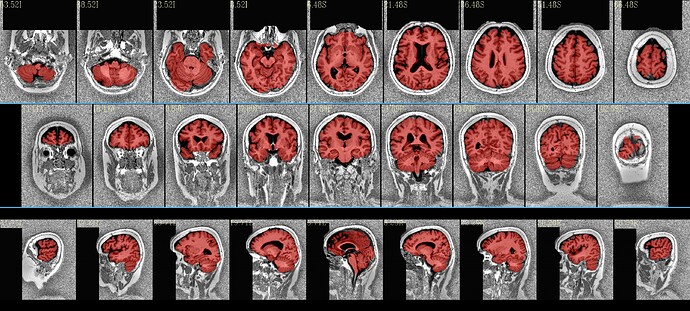
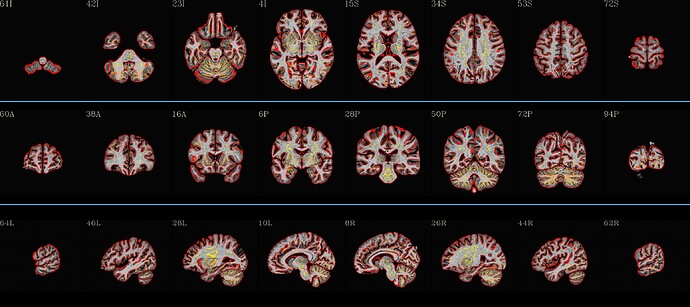
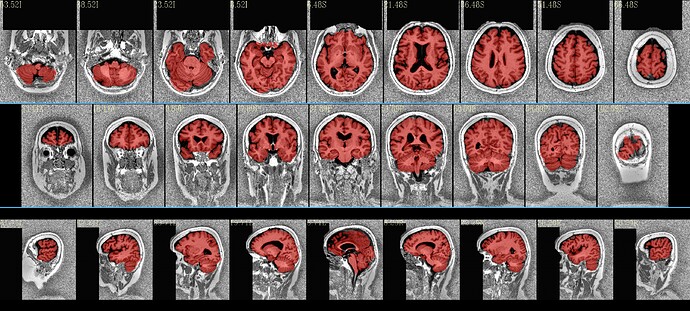
I have recently looked at the MPI-Lemon data a bit for processing. Here is an example of combined skullstripping (SS) and nonlinear warping to a template (the MNI 152) for their sub-032301, using AFNI’s @SSwarper. Because this is a nonstandard T1w volume, as you have pointed out (elevated background noise outside the brain), I tried a few different things. The @SSwarper tool creates automatic QC images, and I will show one of where the brain boundary is found for each case.
NB: in each case below, I am using tcsh syntax for a couple variables and to capture terminal text output in a log file; but those parts can be turned into any kind of scripting you prefer.
Case 1: default options
set subj = sub-032301
set anat = ${subj}_ses-01_acq-mp2rage_T1w.nii.gz
time @SSwarper \
-input ${anat} \
-base MNI152_2009_template_SSW.nii.gz \
-subid ${subj} \
-odir aw_${subj} \
|& tee log_aw_${subj}
… which produces pretty good results:
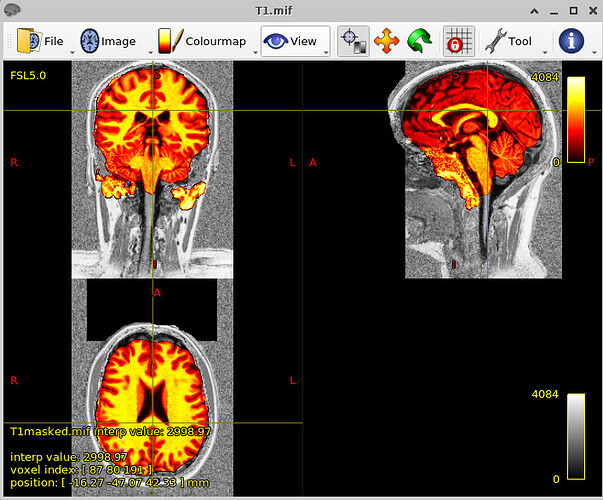
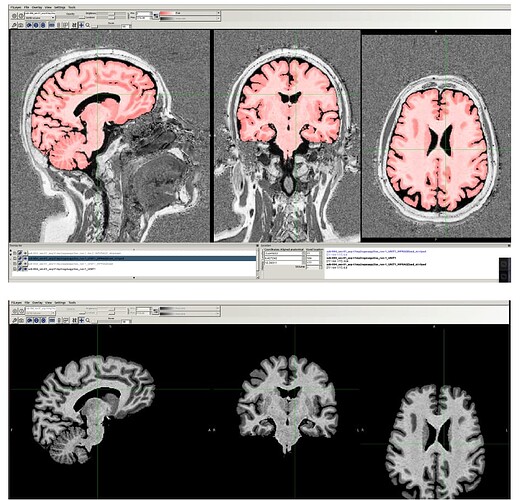
… and just to give a sense of the alignment associated with this—for this run, the following image shows the anatomical warped to the chosen standard space, with the edges of the MNI template overlaid (also pretty good):
If the skullstripping is good, typically the alignment is good, and vice versa, because this program is toggling back and forth between both operations to improve both.
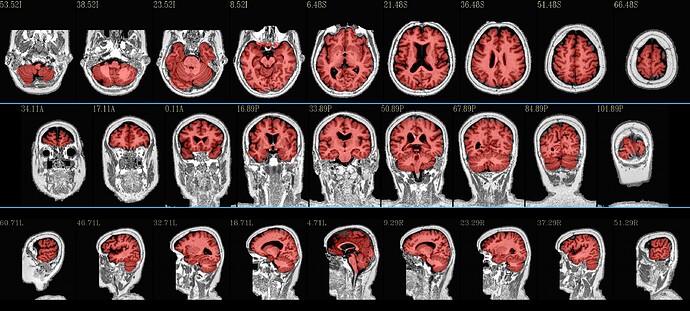
Case 2: put in an opt to turn off initial skull stripping in the program, because the high-background makes this not very good (note how the default above still did quite well, even so, by the end):
set subj = sub-032301
set anat = ${subj}_ses-01_acq-mp2rage_T1w.nii.gz
time @SSwarper \
-input ${anat} \
-base MNI152_2009_template_SSW.nii.gz \
-subid ${subj} \
-odir aw2_${subj} \
-init_skullstr_off \
|& tee log_aw2_${subj}
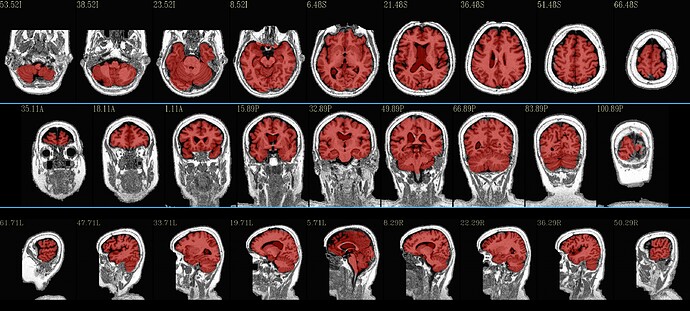
Case 4 (yes, #3 wasn’t very useful in output): use an accompanying dataset in the MPI-Lemon anat/ folder to punch away some of the background—specfically the “*inv-2_mp2rage.nii.gz” dset, which itself has a relatively low background. This is done with automasking using a very low clipping fraction, to preserve as much brain as possible there (some of these dsets also have brightness inhomogeneity). Then use that automasking to punch away background (hopefully) from the T1w volume using 3dcalc:
set subj = sub-032301
set anat = ${subj}_ses-01_acq-mp2rage_T1w.nii.gz
3dAutomask \
-overwrite \
-prefix big_mask.nii \
-clfrac 0.1 \
sub-032301_ses-01_inv-2_mp2rage.nii.gz
3dcalc \
-overwrite \
-a big_mask.nii \
-b ${anat} \
-expr 'step(a)*b' \
-prefix anat_masked.nii.gz
time @SSwarper \
-input anat_masked.nii.gz \
-base MNI152_2009_template_SSW.nii.gz \
-subid ${subj} \
-odir aw4_${subj} \
#-unifize_off \
-SSopt '-blur_fwhm 2' \
|& tee log_aw4_${subj}
Case 5: employing similar, loose “pre-masking” of background, with adding in a couple options to @SSwarper:
set subj = sub-032301
set anat = ${subj}_ses-01_acq-mp2rage_T1w.nii.gz
3dAutomask \
-overwrite \
-prefix big_mask.nii \
-clfrac 0.1 \
sub-032301_ses-01_inv-2_mp2rage.nii.gz
3dcalc \
-overwrite \
-a big_mask.nii \
-b ${anat} \
-expr 'step(a)*b' \
-prefix anat_masked.nii.gz
time @SSwarper \
-input anat_masked.nii.gz \
-base MNI152_2009_template_SSW.nii.gz \
-subid ${subj} \
-odir aw5_${subj} \
-unifize_off \
-SSopt '-blur_fwhm 2' \
|& tee log_aw5_${subj}
Most of these results seem good-to-quite-good, I think. There are little blips around in each case, perhaps, but overall they seem to do a good job. Plus, you get alignment to standard space calculated simultaneously!
–pt