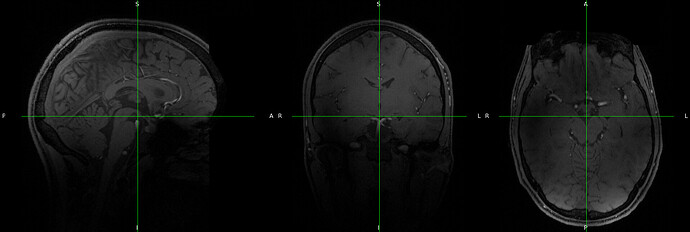
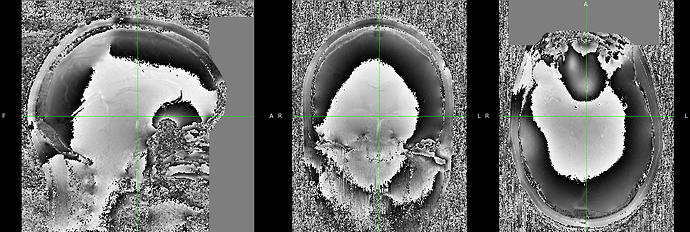
Series_008_[MR_-_ Series_008_[MR_-_tfl_wip944_b17stx_NEW_MP2RAGE_INV2]_tf1_wip944_b17stx_NEW_MP2RAGE_INV2_19700101000000_8.json :
{
"Modality": "MR",
"MagneticFieldStrength": 6.98,
"ImagingFrequency": 297.217,
"Manufacturer": "Siemens",
"ManufacturersModelName": "DicomCleaner",
"DeviceSerialNumber": "18973",
"StationName": "PSYCH15-015_1111",
"PatientPosition": "HFS",
"SoftwareVersions": "Thu Oct 18 06:41:18 EDT 2018",
"MRAcquisitionType": "3D",
"SeriesDescription": "tfl_wip944_b17stx_NEW_MP2RAGE_INV2",
"ProtocolName": "tfl_wip944_b17stx_NEW_MP2RAGE_INV2",
"ScanningSequence": "GR\\IR",
"SequenceVariant": "SP\\MP",
"ScanOptions": "IR\\PFP",
"SequenceName": "tfl3d1rs",
"ImageType": ["ORIGINAL", "PRIMARY", "M", "ND"],
"SeriesNumber": 8,
"AcquisitionTime": "00:16:9.527000",
"AcquisitionNumber": 1,
"SliceThickness": 0.5,
"SAR": 0.0762767,
"EchoTime": 0.00575,
"RepetitionTime": 5,
"InversionTime": 2.78,
"FlipAngle": 3,
"PercentPhaseFOV": 90.2222,
"PercentSampling": 100,
"PhaseEncodingSteps": 305,
"AcquisitionMatrixPE": 406,
"ReconMatrixPE": 406,
"PixelBandwidth": 171,
"ImageOrientationPatientDICOM": [
1,
0,
0,
0,
1,
0 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20211006"
}
Series_009_[MR_-_tfl_wip944_b17stx_NEW_MP2RAGE_INV2]_tfl_wip944_b17stx_NEW_MP2RAGE_INV2_19700101000000_9.json :
{
"Modality": "MR",
"MagneticFieldStrength": 6.98,
"ImagingFrequency": 297.217,
"Manufacturer": "Siemens",
"ManufacturersModelName": "DicomCleaner",
"DeviceSerialNumber": "18973",
"StationName": "PSYCH15-015_1111",
"PatientPosition": "HFS",
"SoftwareVersions": "Thu Oct 18 06:41:18 EDT 2018",
"MRAcquisitionType": "3D",
"SeriesDescription": "tfl_wip944_b17stx_NEW_MP2RAGE_INV2",
"ProtocolName": "tfl_wip944_b17stx_NEW_MP2RAGE_INV2",
"ScanningSequence": "GR\\IR",
"SequenceVariant": "SP\\MP",
"ScanOptions": "IR\\PFP",
"SequenceName": "tfl3d1rs",
"ImageType": ["ORIGINAL", "PRIMARY", "M", "ND"],
"SeriesNumber": 9,
"AcquisitionTime": "00:08:14.822000",
"AcquisitionNumber": 1,
"SliceThickness": 0.5,
"SAR": 0.0994492,
"EchoTime": 0.00575,
"RepetitionTime": 5,
"InversionTime": 2.78,
"FlipAngle": 3,
"PercentPhaseFOV": 90.2222,
"PercentSampling": 100,
"PhaseEncodingSteps": 305,
"AcquisitionMatrixPE": 406,
"ReconMatrixPE": 406,
"PixelBandwidth": 171,
"ImageOrientationPatientDICOM": [
1,
0,
0,
0,
0.987136,
-0.159881 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20211006"
}
Series_007_[MR_-_tf1_wip944_b17stx_NEW_MP2RAGE_UNI_Images]_tf1_wip944_b17stX_NEW_MP2RAGE_UNI_Images_19700101000000_7.json :
{
"Modality": "MR",
"MagneticFieldStrength": 6.98,
"ImagingFrequency": 297.217,
"Manufacturer": "Siemens",
"ManufacturersModelName": "DicomCleaner",
"DeviceSerialNumber": "18973",
"StationName": "PSYCH15-015_1111",
"PatientPosition": "HFS",
"SoftwareVersions": "Thu Oct 18 06:41:18 EDT 2018",
"MRAcquisitionType": "3D",
"SeriesDescription": "tfl_wip944_b17stx_NEW_MP2RAGE_UNI_Images",
"ProtocolName": "tfl_wip944_b17stx_NEW_MP2RAGE_UNI_Images",
"ScanningSequence": "GR\\IR",
"SequenceVariant": "SP\\MP",
"ScanOptions": "IR\\PFP",
"SequenceName": "tfl3d1rs",
"ImageType": ["ORIGINAL", "PRIMARY", "M", "ND", "UNI"],
"SeriesNumber": 7,
"AcquisitionTime": "00:08:47.383000",
"AcquisitionNumber": 1,
"SliceThickness": 0.5,
"SAR": 0.0983129,
"EchoTime": 0.00575,
"RepetitionTime": 5,
"PercentPhaseFOV": 90.2222,
"PercentSampling": 100,
"PhaseEncodingSteps": 305,
"AcquisitionMatrixPE": 406,
"ReconMatrixPE": 406,
"PixelBandwidth": 171,
"ImageOrientationPatientDICOM": [
1,
0,
0,
0,
0.996637,
0.0819385 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
"ConversionSoftwareVersion": "v1.0.20211006"
}
Series_008_[MR_-_tf1_wip944_b17stx_NEW_MP2RAGE_UNI_Images]_tfl_wip944_b17stx_NEW_MP2RAGE_UNI_Images_19700101000000_8.json :
{
"Modality": "MR",
"MagneticFieldStrength": 6.98,
"ImagingFrequency": 297.217,
"Manufacturer": "Siemens",
"ManufacturersModelName": "DicomCleaner",
"DeviceSerialNumber": "18973",
"StationName": "PSYCH15-015_1111",
"PatientPosition": "HFS",
"SoftwareVersions": "Thu Oct 18 06:41:18 EDT 2018",
"MRAcquisitionType": "3D",
"SeriesDescription": "tfl_wip944_b17stx_NEW_MP2RAGE_UNI_Images",
"ProtocolName": "tfl_wip944_b17stx_NEW_MP2RAGE_UNI_Images",
"ScanningSequence": "GR\\IR",
"SequenceVariant": "SP\\MP",
"ScanOptions": "IR\\PFP",
"SequenceName": "tfl3d1rs",
"ImageType": ["ORIGINAL", "PRIMARY", "M", "ND", "UNI"],
"SeriesNumber": 8,
"AcquisitionTime": "00:08:12.935000",
"AcquisitionNumber": 1,
"SliceThickness": 0.5,
"SAR": 0.0994492,
"EchoTime": 0.00575,
"RepetitionTime": 5,
"PercentPhaseFOV": 90.2222,
"PercentSampling": 100,
"PhaseEncodingSteps": 305,
"AcquisitionMatrixPE": 406,
"ReconMatrixPE": 406,
"PixelBandwidth": 171,
"ImageOrientationPatientDICOM": [
1,
0,
0,
0,
0.987136,
-0.159881 ],
"InPlanePhaseEncodingDirectionDICOM": "COL",
"ConversionSoftware": "dcm2niix",
Thanks @jsein !!