I am working with EEG but I have associated MRI in Analyze format. I have converted them to NIFTI format. However, the BIDS validator returns an error because it says the dimension is 4 instead of 3. Yet, even if the dimension is 4, it seems that the 4th dimension is not used. How can I remove the 4th dimension on the NIFTI file so it passes BIDS validation?
This is a link to the NIFTI and the original Analyze files.
Arno
/fslinfo s01.nii
data_type INT16
dim1 256
dim2 256
dim3 256
dim4 1
datatype 4
pixdim1 1.000000
pixdim2 1.000000
pixdim3 1.000000
pixdim4 0.000000
cal_max 0.000000
cal_min 0.000000
file_type NIFTI-1+
Arno-
I suspect the issue is that the tool used to convert the Analyze images to NIfTI did not correctly set dim0. This should be the number of dimensions. In your case, this should be 3, but I suspect that if you look at your header with fslhd you will see that it has set this to 4. So update your conversion tool to correctly specify dim0.
fslhd s01.nii
....
dim0 4
dim1 256
dim2 256
dim3 256
dim4 1
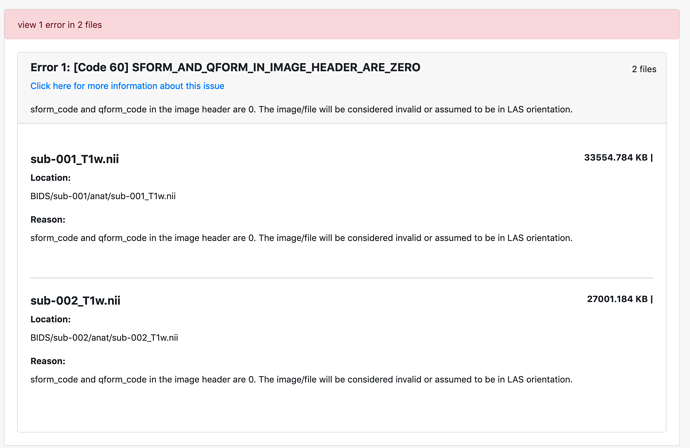
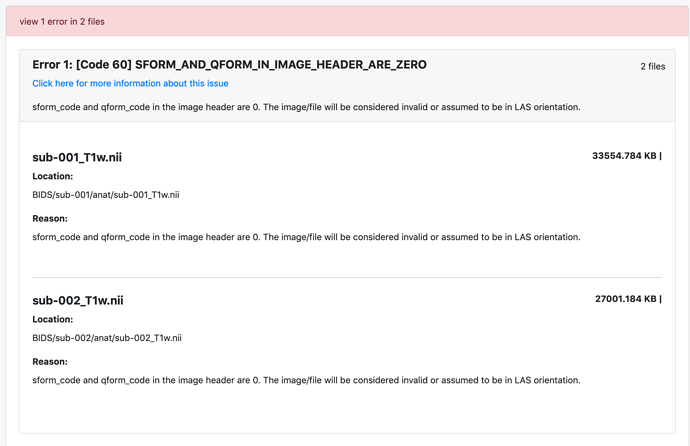
I have passed the first hurdle by using different converter but now get this error about orientation. Any pointer would help.
Arno
The Analyze header did not provide spatial orientation information. This is a major difference between NIfTI and Analyze. I would strongly recommend converting images directly from the scanner to NIfTI format using a tool like dcm2niix that will preserve the spatial orientation. Failing that, if you have Matlab, I would use SPM12 to first “Check reg” to approximately set the rotations and ensure the origin is near the anterior commissure. Ideally you would run SPM’s “Coregister” function to provide a better starting estimate (just ‘estimate’ to have it set rotations and origin without reslicing the data). If you do not have Matlab, use FSLeyes or MRIcroGL to set the rotation (in MRIcroGL, Tools/Rotation).
Given that the brain is roughly symmetric on the left-right axis, you will want to make sure the images are not flipped along this axis. From my perspective, Analyze has not been a frontline tool since 2002, so you may want to check if other tools can be updated to preserve spatial information.
Thank you Chris. Using MRIcroGL does allow to set the orientation and when I re-open the file in MRIcroGL, I can see that the axes are labeled (instead of ?). However, it still does not pass the validator with an error in orientation. I have double checked everything, cleared the cache of my browser (Firefox because for some reason the validator crashes on Chrome - EEG files are too large). Any further hint?
Arno
.
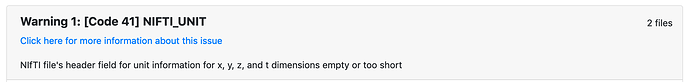
I also get this strange error about units. How can I fix that (searched Neurostars but did not find an obvious response).
Sorry, I have not used the Analyze format since 2002. The easiest thing would be to run SPM12’s ‘Coregister/Estimate’ function. That provides a robust method to estimate the rotations and translations, applying the results in the NIfTI Form/QForm without interpolating the imaging data. Most of the other registration tools want to reslice the data, which would not be ideal (e.g. I think flirt, acpcdetect only work this way). Perhaps ANTS has a good feature for this (it has a very robust initial stage). Sorry, outside my expertise.