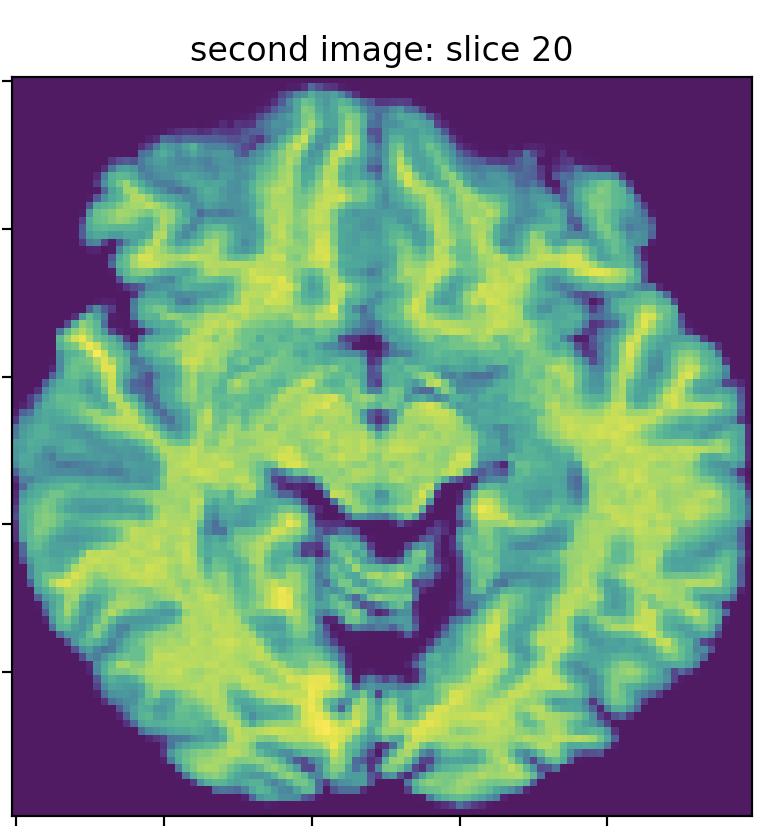
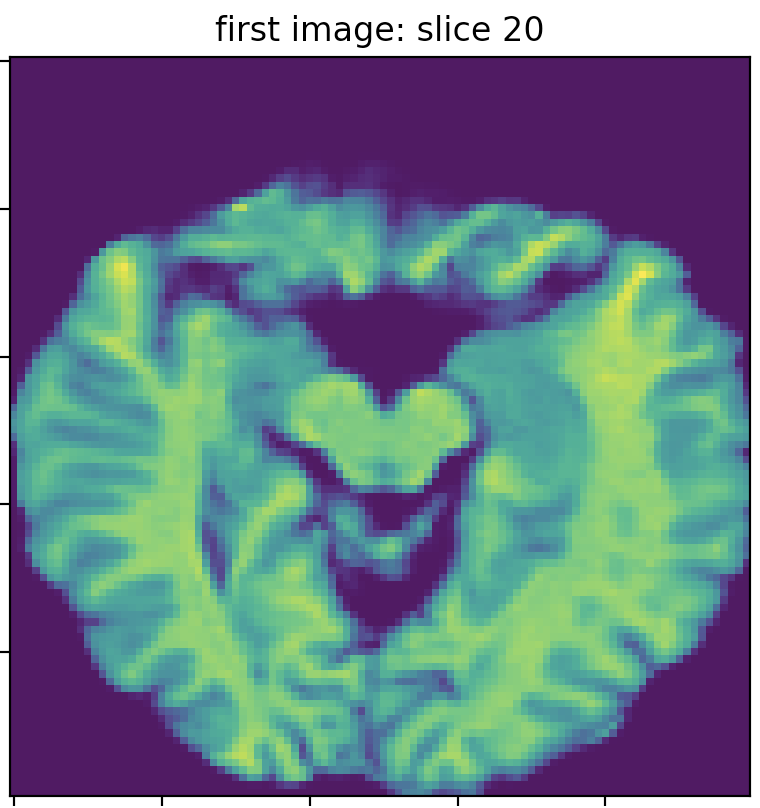
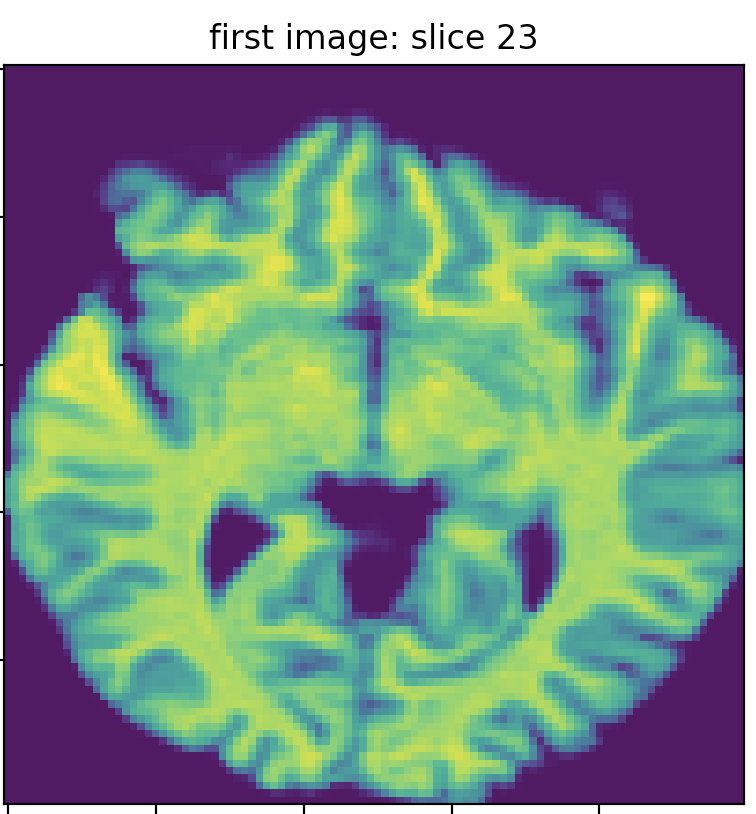
I got two MRI images for the same subject on the same day. The first image’s dim is (142, 352, 352), and the second is (160, 256, 256). I resampled the images to the same dimension (100,100,50) by interpolating the images using `skimage.resize. The following three images show slice 20 of the first and second images, which clearly do not match (in terms of the brain structure/anatomy and not the intensity values). I searched for the best match of the second image slice 20 in the first image slices, and I found the best match is slice 23, which does not precisely fit slice 20 of the second image.
My understanding is that while capturing MRI images, the practitioner calibrates the scanner to capture a specific “slice resolution”. For instance, an MRI image with 100 slices has less axial resolution than another with 300 axial slices. The first MRI image (which has 100 axial slices) has an information loss where some axial slices are skipped and never captured. This is my interpretation of why I am seeing mismatches between two MRI images of the same subject. Is that correct?
Is there a tool compensating for the information loss due to the lower number of slices in MRI images?
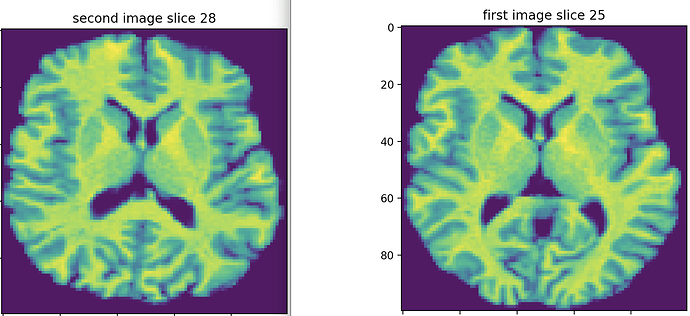
Update (voxels resampling): I found that the new images have different voxel resolutions. One of them has 2x2x2, and the other is 1x1x1 mm. I unified their resolution to 1 mm using the afni package’s program 3dresample. And then, I cropped the empty space using crop_image. Then I resampled the two images to the dimensions to (100,100,50). After that, I found that slice 25 from the first image is the most similar to slice 28 in the second image. They are more similar to each other compared to the ones obtained before voxel size resampling.