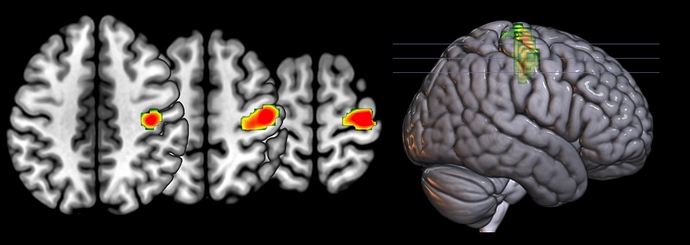
There is a problem with resizing thresholded images: the thresholded images artificially set all non-significant voxels to zero. In reality, since we smooth our images, the non-significant neighbors of significant voxels are almost significant. This means that when we resize thresholded images they the edges appear artificially dark and the size shrinks. There are three solutions:
- Resize the unthresholded image, and then apply thresholding to the upsampled image.
- Use nearest neighbor interpolation. This will create jagged looking images, but does not insert artificially dark voxels.
- Upsize both the unthresholded image and the thresholded image and then use the upsized thresholded image to mask the unthresholded image (see my code below).
Assuming you have the latest release of MRIcroGL (1.2.20190410), you might want to try the Scripting/Templates/Jagged menu item. This will load a low resolution statistical map twice onto a high resolution anatomical scan - once with nearest neighbor interpolation (which creates blocky images) and once with linear interpolation (which creates smoother images but tends to erode thresholded images a bit at the edges). With the graphical interface you can control whether overlays are loaded smooth or not by clicking the “options” button in the layer panel.
The image shows the result of my SPM script (below).
function nii_resizeAndMask(srcNam, mskNam, tarNam)
%Reslice source and mask to match shape of target, then apply mask to source
% Rationale: Masking after reslicing preserves precision
%Inputs
% srcNam: filename for NIfTI source image, e.g. raw t-scores
% mskNam: filename for NIfTI mask image, e.g. cluster thresholded version of source
% tarNam: filename for NIfTI target image: high resolution image used for display
%Examples
% nii_resizeAndMask('spmT_0002.nii', 'k48_T3p12.nii', 'spm152.nii')
if ~exist('srcNam', 'var')
srcNam = spm_select(1,'image','Select source image');
end
if ~exist('mskNam', 'var')
mskNam = spm_select(1,'image','Select mask image');
end
if ~exist('tarNam', 'var')
tarNam = spm_select(1,'image','Select high resolution taget');
end
%load source
shdr = spm_vol(srcNam);
simg = spm_read_vols(shdr);
%load mask:
mhdr = spm_vol(mskNam);
mimg = spm_read_vols(mhdr);
if ~isequal(shdr.dim, mhdr.dim), error('source and mask should have identical resolutions.'); end;
mimg(isnan(mimg)) = 0; %spm uses not-a-number outside brain
mimg = (mimg ~= 0)+0.0; %binarize mask as 0 or 1
%reslice source to target
thdr = spm_vol(tarNam); %load input header
outhdr = shdr;
[~,mnam] = fileparts(mhdr.fname);
[pth,nam,ext] = fileparts(shdr.fname);
outhdr.fname = fullfile(pth,[mnam '_' nam ext]);
outhdr.dim = thdr.dim;
outhdr.mat = thdr.mat;
outimg = zeros(outhdr.dim(1:3));
for i = 1:outhdr.dim(3)
M = inv(spm_matrix([0 0 -i])*inv(outhdr.mat)*shdr.mat);
outimg(:,:,i) = spm_slice_vol(simg, M, outhdr.dim(1:2), 2); % (1=linear interp, 2=spline)
end
%reslice mask
outmimg = zeros(outhdr.dim(1:3));
for i = 1:outhdr.dim(3)
M = inv(spm_matrix([0 0 -i])*inv(outhdr.mat)*mhdr.mat);
outmimg(:,:,i) = spm_slice_vol(mimg, M, outhdr.dim(1:2), 2); % (1=linear interp, 2=spline)
end
%threshold mask 0.5 - 50%
outmimg = (outmimg >= 0.5)+0.0; %binarize mask as 0 or 1
outimg(outmimg == 0) = NaN;
%save result
spm_write_vol(outhdr,outimg);