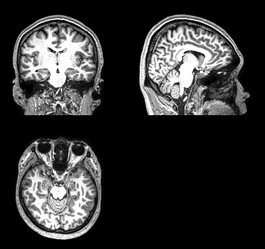
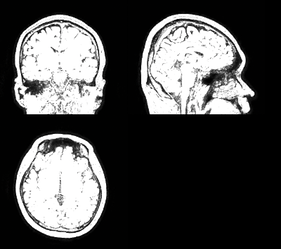
Summary of what happened:
I’m processing a dataset with around 160 participants. For a few participants, MRIQC crashes when calculating the quality metrics for the T1-weighted image. I cannot see what’s wrong with those images. They also run through fMRIPrep without a problem.
Command used:
docker run --rm -v ${infolder}:/data:ro \
-v ${outfolder}:/out \
nipreps/mriqc:latest /data /out participant \
--participant_label ${subject} \
--nprocs 16
Version:
docker image: 936fc4d9cfbd
Data formatted according to a validatable standard? Please provide the output of the validator:
bids-validator@1.14.1
This dataset appears to be BIDS compatible.
Summary: Available Tasks: Available Modalities:
4174 Files, 119.17GB Raven Progressive Matrices MRI
167 - Subjects
1 - Session
Relevant log outputs (up to 20 lines):
2024-12-16 15:02:13 |e[31;20m ERROR e[0m|e[31;20m nipype.workflow e[0m|e[31;20m Node ComputeQI2.a0 (taskid=155) crashed: /out/logs/crash-20241216-150213-root-ComputeQI2.a0-860b054d-6194-48fe-b2f9-8c665c2d7c3c.txte[0m
Traceback (most recent call last):
File "/opt/conda/bin/mriqc", line 8, in <module>
sys.exit(main())
^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/cli/run.py", line 178, in main
mriqc_wf.run(**_plugin)
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 196, in run
notrun.append(self._clean_queue(jobid, graph, result=result))
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 259, in _clean_queue
raise RuntimeError(''.join(result['traceback']))
RuntimeError: Traceback (most recent call last):
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 64, in run_node
result['result'] = node.run(updatehash=updatehash)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node ComputeQI2.
Traceback:
Traceback (most recent call last):
File "/opt/conda/lib/python3.11/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/interfaces/anatomical.py", line 378, in _run_interface
qi2, out_file = art_qi2(imdata, airdata)
^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/qc/anatomical.py", line 497, in art_qi2
kde_skl = KernelDensity(kernel='gaussian', bandwidth=4.0).fit(modelx[:, np.newaxis])
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/sklearn/base.py", line 1351, in wrapper
return fit_method(estimator, *args, **kwargs)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/sklearn/neighbors/_kde.py", line 226, in fit
X = self._validate_data(X, order="C", dtype=np.float64)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/sklearn/base.py", line 633, in _validate_data
out = check_array(X, input_name="X", **check_params)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/sklearn/utils/validation.py", line 1003, in check_array
_assert_all_finite(
File "/opt/conda/lib/python3.11/site-packages/sklearn/utils/validation.py", line 126, in _assert_all_finite
_assert_all_finite_element_wise(
File "/opt/conda/lib/python3.11/site-packages/sklearn/utils/validation.py", line 175, in _assert_all_finite_element_wise
raise ValueError(msg_err)
ValueError: Input X contains NaN.
KernelDensity does not accept missing values encoded as NaN natively. For supervised learning, you might want to consider sklearn.ensemble.HistGradientBoostingClassifier and Regressor which accept missing values encoded as NaNs natively. Alternatively, it is possible to preprocess the data, for instance by using an imputer transformer in a pipeline or drop samples with missing values. See https://scikit-learn.org/stable/modules/impute.html You can find a list of all estimators that handle NaN values at the following page: https://scikit-learn.org/stable/modules/impute.html#estimators-that-handle-nan-values
Relevant information:
I checked the actual image for NaN values and did not find any.