Tajwar
July 11, 2020, 11:37am
1
Hello. I ran MRIQC docker command:
docker run -it --rm -v /c/Users/Username/Documents/BIDS_data:/data:ro -v /c/Users/Username/Documents/mriqc-output:/out poldracklab/mriqc:latest /data /out participant --participant_label 008 --no-sub
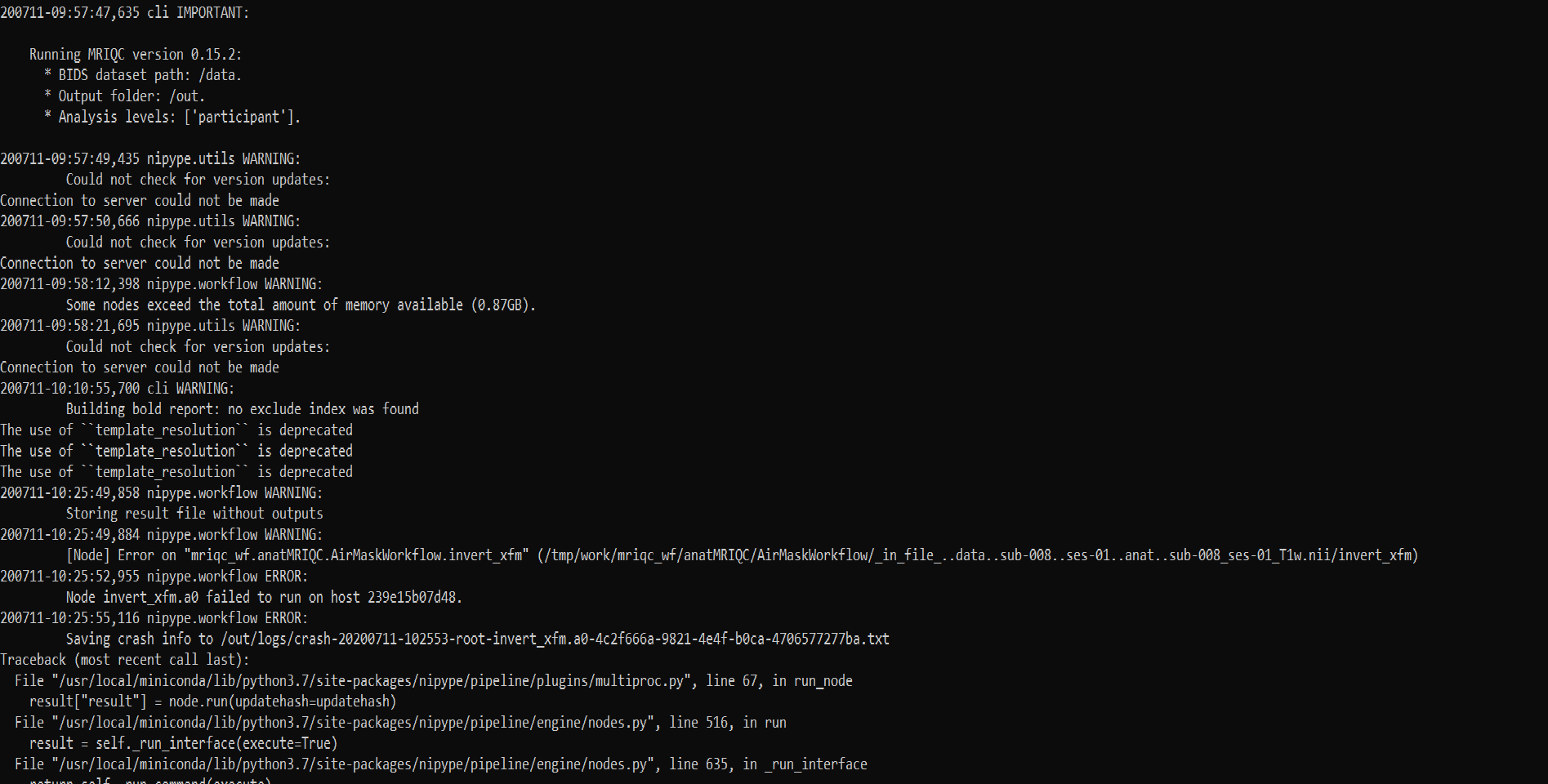
It terminated in between giving runtime error code 137.
I am using docker toolbox on WSL1 (Cannot upgrade to WSL2 because windows updates 2004 has not reached to my system yet). As I found out about error code 137, it is related to RAM limit for docker. Since I am using docker toolbox, the only option to change docker settings is through VM virtualbox and I was unable to find any option to change memory settings there.
Is there any way that I can use mriqc and get results?
Thanks
Hi @Tajwar seems like it is caused by memory starvation. The usual recommendation is to use Docker Desktop instead of Docker Toolbox, but since you cannot update as mentioned, you could try out the following:
Step 1 : Remove the existing Docker VM
docker-machine rm default
Step 2 : Create a new VM with an increased amount of RAM (Depends on the RAM available on your PC). In this sample command, I will include 4GB RAM:
docker-machine create -d virtualbox --virtualbox-cpu-count=2 --virtualbox-memory=4096 --virtualbox-disk-size=50000 default
Now restart Docker and run the command.
Tajwar
July 12, 2020, 5:22pm
3
Thanks @sanjula . It solved the problem.
1 Like
Great! Happy to know it got solved. Let us know if there are other queries, the community is always there to help!
Dear experts,
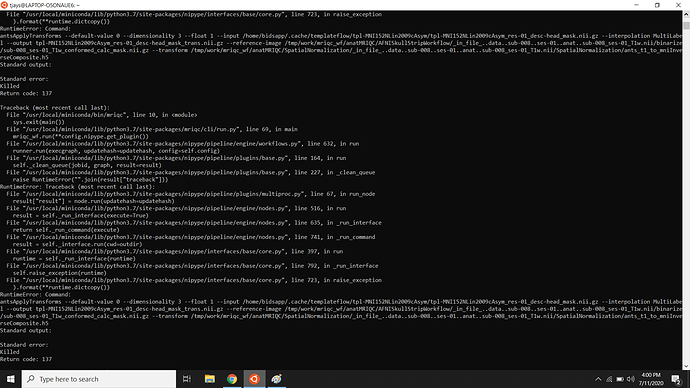
it seems I am having the same error when running the MRIQC docker in WSL (in Windows 11) - see the full bash input and output below.
I already tried to give WSL as much memory as possible, but no luck so far…
Thank you!
sudo docker run -it --rm \
-v /mnt/c/Users/marco/HiDrive/KKB/QA/dcm2bids_mriqc_pipeline/dcm2bids_out/:/data:ro \
-v /mnt/c/Users/marco/HiDrive/KKB/QA/dcm2bids_mriqc_pipeline/mriqc_out/:/out \
nipreps/mriqc:latest \
/data /out participant \
--no-sub
------------------------------------------------------------------
Running MRIQC version 24.1.0.dev0+g3fe90466.d20240417
----------------------------------------------------------------
* BIDS dataset path: /data.
* Output folder: /out.
* Analysis levels: ['participant'].
------------------------------------------------------------------
2024-07-19 16:41:25 | IMPORTANT | mriqc | Building MRIQC's workflows...
2024-07-19 16:41:36 | IMPORTANT | mriqc | Workflow building finished (exit code 0).
2024-07-19 16:46:36 | WARNING | nipype.workflow | [Node] Error on "mriqc_wf.anatMRIQC.synthstrip_wf.synthstrip" (/tmp/work/mriqc_wf/anatMRIQC/synthstrip_wf/_in_file_..data..sub-001..ses-001..anat..sub-001_ses-001_T1w.nii.gz/synthstrip)
2024-07-19 16:46:36 | ERROR | nipype.workflow | Node synthstrip.a0 (taskid=16) crashed: /out/logs/crash-20240719-164636-root-synthstrip.a0-4a037e74-1d8a-485c-87d5-4cda31526432.txt
Traceback (most recent call last):
File "/opt/conda/bin/mriqc", line 8, in <module>
sys.exit(main())
^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/cli/run.py", line 178, in main
mriqc_wf.run(**_plugin)
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 196, in run
notrun.append(self._clean_queue(jobid, graph, result=result))
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 259, in _clean_queue
raise RuntimeError(''.join(result['traceback']))
RuntimeError: Traceback (most recent call last):
File "/opt/conda/lib/python3.11/site-packages/mriqc/engine/plugin.py", line 64, in run_node
result['result'] = node.run(updatehash=updatehash)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
^^^^^^^^^^^^^^^^^^^^^^^^^^
File "/opt/conda/lib/python3.11/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node synthstrip.
Cmdline:
synthstrip -b 1 -i /tmp/work/mriqc_wf/anatMRIQC/synthstrip_wf/_in_file_..data..sub-001..ses-001..anat..sub-001_ses-001_T1w.nii.gz/pre_n4/clipped_corrected.nii.gz --model /opt/freesurfer/models/synthstrip.1.pt -n 1 -o clipped_corrected_desc-brain.nii.gz -m clipped_corrected_desc-brain_mask.nii.gz
Stdout:
Stderr:
Killed
Traceback:
RuntimeError: subprocess exited with code 137.
Steven
July 19, 2024, 4:57pm
6
Hi @marco5790 ,
That indicates you are using insufficient RAM for the job. What are you system specs and how much data are you processing?
Best,
Hey Steven,
at this point just one test subject with one bold dataset (200mb) and a T1 (12mb).
Unfortunately WSL is very limited in the resources it has available:
Best,
Steven
July 19, 2024, 5:07pm
8
You might instead consider brainlife.io or other alternatives for cloud-based neuroimaging preprocessing. I don’t think trying to work with a limited resource is sustainable.
Ok I will move to a proper server then. Thank you!