Hi all,
I’m currently working with 400+ T1w images that I downloaded from an open-access database. Given that I didn’t collect the images myself and therefore can’t be sure of their quality, I decided to try MRIQC for the first time.
MRIQC (version: 0.15.2rc1) ran on the HPC cluster without issue, but when I checked the individual anatomical reports, I noticed two warning messages under “Workflow details”:
- Detected hat mask was too small
- Detected a zero-filled frame, has the image been rotated?
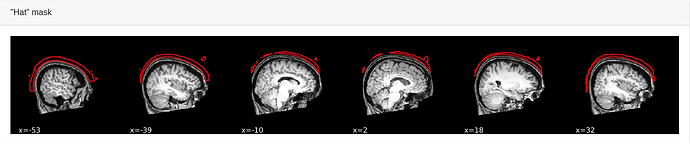
I couldn’t visually identify any obvious errors in the images I reviewed, so I re-ran MRIQC with the “–verbose-reports” argument. Below is an image of the “hat” mask. It looks quite different to the one shown in the example provided in the MRIQC documentation and others I’ve checked on OpenNeuro:
Based on the above, I want to know the following:
- What is likely to be causing the warnings?
- Do the warnings indicate the presence of issues that might have affected the IQMs derived by MRIQC?
- If so, how do I address these issues?
Any help would be very much appreciated! I’ve spent far too long searching Google for solutions…
Cheers,
Rikki
I have the same warning.
Did you identify a solution (and cause)?
Thanks.
-Tom.
I’m afraid not. I ended up opting for a different QC method, as I didn’t have a firm understanding of the issue or its cause at the time. I’m aware of at least one other person who has encountered the same warning: https://groups.google.com/g/mriqc-users/c/9GQ1ajllvaw/m/7amXlPLQAgAJ?utm_medium=email&utm_source=footer
From my reading of the documentation, the error seems to arise from step 5 of the anatomical workflow (described here). This step is conducted by the Python function airmsk_wf(), which carries out background region segmentation (for more info, see here).
Looking at the source code, it seems that the “hat mask” warning is generated when the calculated air mask is smaller than 5e-5. I’m guessing that this means there’s insufficient background (i.e. air) in the structural MR images being preprocessed. I really need to take a deeper look…
Sorry I can’t provide any more info at this stage! Let me know if you work it out.
Thanks, your insights are very much appreciated.
It seemed my pipeline was picking up the wrong file.
When the correct T1 was used the issue resolved.