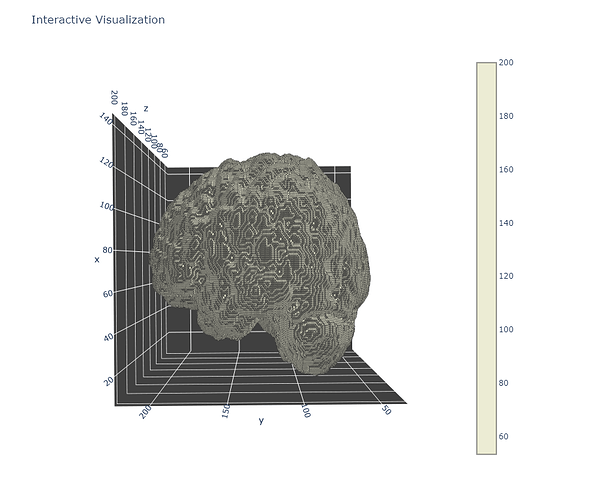
Background: I am trying to generate mesh of lesion segmentation from NIFTI file, so I am succesfully generated it with measure.marching_cubes_lewiner, in advance transformed NIFTI to ndarray.
However, i tried to check the orientation of the mesh, and found out that it is wrong - z coordinate is stretching the brain mesh
I consider that the problem is in nibabel orientation, because while loading images with nib.load:
def show_slices(stack):
fig, axes = plt.subplots(1, len(stack))
for i, slice in enumerate(stack):
axes[i].imshow(slice, cmap="gray")
plt.show()
sample_stack(array_img, brain_mask_img)
i have following orientation:
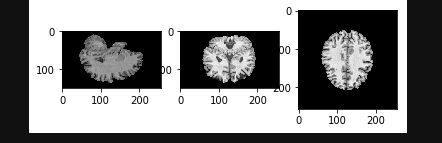
How to resolve issue with such orientation and allign it with FOV window, so all coordinates wouldn’t stretch meshes?