A colleague will be sending me a series of nifti files converted with dcm2niix, along with their .json side-cars. These would be single participant, single session. Does there exist a tool that organizes nifti+json pairs into a bids structure, or would this be something I simply need to do manually?
From a colleague in Paris:
Here is a list of software available for BIDS conversion : https://bids.neuroimaging.io/benefits#mri-and-pet-converters
https://github.com/benoitberanger/niix2bids has different objective : it is made for fast BIDS conversion but has to cut some corners. And It only works for Siemens magnets for now. I made this tool in order for our lab to convert lors of old datasets without any configuration config file requirement.
For one-offs or small datasets I usually do the conversion semi-manually, with an R script like below. Part of why I prefer this manual style is to be able to double-check the conversion (having an explicit record of which scans were assigned which names can be invaluable). Some programs create records along these lines, but for me, the time needed to configure and verify the programs can be longer than manual scripting for small datasets.
Here’s a bit of a script to give the idea (an old, but complete script is here; I can share updated versions if there’s interest):
sub.id <- "9998"; # subject ID
in.path <- paste0("C:/BIDS/dicoms/", sub.id, "/scans/"); # top-level path to source dicoms
ds.path <- paste0("C:/BIDS/bids/sub-", sub.id, "/ses-1/"); # top-level path to where session will be written in BIDS format
dcm.path <- "C:/Users//MRIcroGL_windows/MRIcroGL/Resources/dcm2niix.exe"; # https://www.nitrc.org/projects/mricrogl/
# function to rename the created .nii.gz and .json from the name given them by dcm2niix to proper BIDS.
# The function prints TRUE if everything worked properly, an error message if not.
# img.num <- 9; # first number in the dicom scans directory name, e.g., 9_T1 MPRAGE_sag_p2 iso 1_176
# new.fname <- paste0("sub-", sub.id, "_ses-1_T1w"); # desired BIDS name for this file
do.rename <- function(img.num, new.fname) {
file.rename(from=list.files(out.path, pattern=paste0("_", img.num, ".json"), full.names=TRUE), to=paste0(out.path, new.fname, ".json"));
file.rename(from=list.files(out.path, pattern=paste0("_", img.num, ".nii.gz"), full.names=TRUE), to=paste0(out.path, new.fname, ".nii.gz"));
}
####### anat (T1 and T2 anatomy)
out.path <- paste0(ds.path, "anat/");
if (!dir.exists(out.path)) { dir.create(out.path); }
system2(dcm.path, args=paste0("-b y -z y -v y -o ", out.path, ' -f "%t_%p_%s" "', in.path, '23_T1 MPRAGE_sag_p2 iso 1_176"'), stdout=TRUE);
do.rename(23, paste0("sub-", sub.id, "_ses-1_T1w"));
system2(dcm.path, args=paste0("-b y -z y -v y -o ", out.path, ' -f "%t_%p_%s" "', in.path, '24_t2_spc_sag_p2_iso 1"'), stdout=TRUE);
do.rename(24, paste0("sub-", sub.id, "_ses-1_T2w"));
####### func (bold & sbref)
out.path <- paste0(ds.path, "func/");
if (!dir.exists(out.path)) { dir.create(out.path); }
system2(dcm.path, args=paste0("-b y -z y -v y -o ", out.path, ' -f "%t_%p_%s" "', in.path, '11_tfMRI_IAPS1_3p0FA66_AP_SBRef"'), stdout=TRUE);
do.rename(11, paste0("sub-", sub.id, "_ses-1_task-IAPS_run-1_sbref"));
system2(dcm.path, args=paste0("-b y -z y -v y -o ", out.path, ' -f "%t_%p_%s" "', in.path, '12_tfMRI_IAPS1_3p0FA66_AP"'), stdout=TRUE);
do.rename(12, paste0("sub-", sub.id, "_ses-1_task-IAPS_run-1_bold"));
# etc.
@benoitberanger the dcm2niix source code contains some hints on deriving modality from a Siemens MRI using the DICOM sequence name (0018,0024) which is stored as SequenceName in the BIDS JSON:
- ep_b: dwi
- epfid2d: perf
- epfid2d: bold
- epfid3d1_15: swi
- epse2d: dwi (when b-vals specified)
- epse2d: fmap (spin echo, e.g. TOPUP, nb could also be extra B=0 for DWI sequence)
- fl2d: localizer
- fl3d1r_t: angio
- fl3d1r_tm: angio
- fl3d1r: angio
- fl3d1r: swi
- fl3d1r: ToF
- fm2d: fmap (gradient echo, e.g. FUGUE)
- spc3d: T2
- spcir: flair (dark fluid)
- spcR: PD
- tfl3d: T1
- tfl_me3d5_16ns: T1 (ME-MPRAGE)
- tir2d: flair
- tse2d: PD (short TE)
- tse2d: T2 (long TE)
- tse3d: T2
@neurolabusc thanks, this list will be useful for the decision tree niix2bids/config_file/siemens.py
@Remi-Gau The converter is not tested intensively. Is it ok to wait before adding it on the website ? In the next weeks/months I should have access to some large datasets from other MRI labs. This will help me check the robustness of the code.
@benoitberanger I have archived Siemens data that you might find useful. Most of my archives are designed to examine specific issues rather than typical datasets. However, you may find them useful. You can look at my OSF datasets, archival data, and many of my dcm_qa validation datasets e.g. dcm_qa_fmap, dcm_qa_stc, dcm_qa_nih, dcm_qa, dcm_qa_asl, etc.
oh yeah of course.
just ping me when you feel OK having it listed there.
The nibabel plugin from BIDScoin can (easily) convert nifti datasets to BIDS:
https://bidscoin.readthedocs.io/en/stable/options.html#nibabel2bids-plugin
Hi, I tried using Nibabel, but I encountered an issue: it cannot automatically generate JSON files. Do you know of any code or application that can generate JSON files from NIfTI automatically? Thanks!
Hi @ShuningW and welcome to neurostars!
JSONs contains information not found in NIFTI files, so no software can create the jsons from nifti. You will have to use information from the scanning protocol, or a Dicom using the same sequence in another dataset if you have that available.
The minimal set of fields tools like fmriprep use are pretty small. I think they are (for BOLD files):
"RepetitionTime" : # You can probably get this from the spacing in 4th dimension revealed by 'fslinfo'
"TotalReadoutTime" :
"PhaseEncodingDirection":
"TaskName": # don't need scanner protocol for that, you just put the name in
"SliceTiming": # not required but recommended
"B0FieldSource": # if using B0FieldIdentifier in fmaps
for fmaps, these may be
"TotalReadoutTime" :
"PhaseEncodingDirection": # if using RPE fieldmap
"Echo1": # if using GRE fieldmap
"Echo2":# if using GRE fieldmap
"B0FieldIdentifier": # If not using IntendedFor
"IntendedFor" # If not using B0FieldIdentifier/Source
T1w does not have required fields.
If parameters are the same across all BOLD / fmap files, then you can please these files in the BIDS root directory and they will be inherited to all subjects.
Best,
Steven
Hi Steven,
Thank you very much for your help!
I am working on converting NIfTI files to BIDS format and subsequently using fMRIPrep. However, I currently do not have any DICOM data available.
Could you please let me know if it is possible to meet the necessary requirements solely by extracting information from the NIfTI headers? There are many numbers in the header, and I am a bit uncertain about the meaning of some of them.
<class 'nibabel.nifti1.Nifti1Header'> object, endian='<'
sizeof_hdr : 348
data_type : np.bytes_(b'')
db_name : np.bytes_(b'')
extents : 0
session_error : 0
regular : np.bytes_(b'r')
dim_info : 48
dim : [ 4 64 64 41 167 1 1 1]
intent_p1 : 0.0
intent_p2 : 0.0
intent_p3 : 0.0
intent_code : none
datatype : int16
bitpix : 16
slice_start : 0
pixdim : [-1.0000000e+00 3.0000000e+00 3.0000000e+00 3.0000000e+00
2.0000000e+00 1.0000000e+00 2.4499811e-01 5.1467859e+04]
vox_offset : 0.0
scl_slope : nan
scl_inter : nan
slice_end : 40
slice_code : alternating increasing
xyzt_units : 10
cal_max : 0.0
cal_min : 0.0
slice_duration : 0.0
toffset : 0.0
glmax : 255
glmin : 0
descrip : np.bytes_(b'TE=25;sec=51467.8600;phaseDir=+;dwell=0.245')
aux_file : np.bytes_(b'')
qform_code : scanner
sform_code : scanner
quatern_b : -0.016290896
quatern_c : 0.9998301
quatern_d : 0.0032194268
qoffset_x : 102.17818
qoffset_y : -94.751114
qoffset_z : -29.85758
srow_x : [-2.9980240e+00 -9.7883224e-02 -4.7655471e-02 1.0217818e+02]
srow_y : [-9.7574309e-02 2.9983454e+00 -2.0094888e-02 -9.4751114e+01]
srow_z : [-4.8284844e-02 1.8531669e-02 2.9995542e+00 -2.9857580e+01]
intent_name : np.bytes_(b'')
magic : np.bytes_(b'n+1')
Your guidance on this matter would be greatly appreciated.
Hi @ShuningW,
No, from what I can tell the fields like would need to be found on the scanner protocol sheet or a dataset using a similar / same sequence.
Best,
Steven
Hi, @Steven,
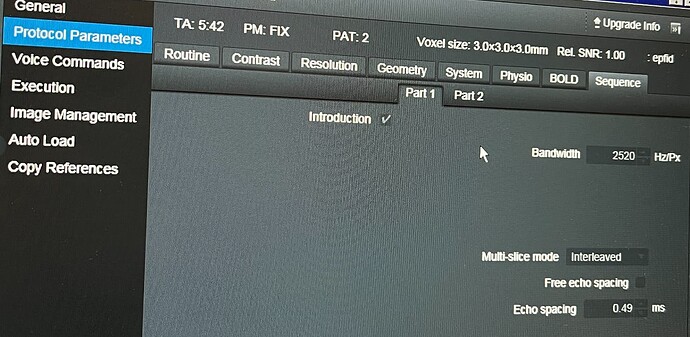
Thank you so much for your help; your answers have been incredibly useful ![]() . Based on your guidance, I reviewed the protocol parameters, but I have some questions regarding the Slice Timing and Total Readout Time.
. Based on your guidance, I reviewed the protocol parameters, but I have some questions regarding the Slice Timing and Total Readout Time.
For Slice Timing, in my project, I know the TR is 2 seconds. A total of 41 axial slices were acquired in an ascending interleaved fashion with no interslice gap to cover the whole brain. So, I divided 2 by 41 to obtain the Slice Timing array, as shown below. Does this look correct?
"SliceTiming": [
0.0, 0.0975609756097561, 0.1951219512195122, 0.2926829268292683, 0.3902439024390244,
0.4878048780487805, 0.5853658536585366, 0.6829268292682927, 0.7804878048780488,
0.8780487804878049, 0.975609756097561, 1.0731707317073171, 1.1707317073170732,
1.2682926829268293, 1.3658536585365855, 1.4634146341463417, 1.5609756097560976,
1.6585365853658536, 1.7560975609756098, 1.853658536585366, 1.951219512195122,
0.04878048780487805, 0.14634146341463414, 0.24390243902439024, 0.34146341463414637,
0.4390243902439024, 0.5365853658536586, 0.6341463414634146, 0.7317073170731707,
0.8292682926829268, 0.9268292682926829, 1.024390243902439, 1.1219512195121952,
1.2195121951219512, 1.3170731707317074, 1.4146341463414633, 1.5121951219512195,
1.6097560975609757, 1.7073170731707317, 1.8048780487804876, 1.9024390243902438
],
Additionally, I couldn’t find the value for Total Readout Time directly in the parameters. We are using a Siemens scanner, and I can only find the Echo Spacing in the interface, as shown in the attached image. Could you please further explain how to determine this value?
TotalReadoutTime can be calculated from EffectiveEchoSpacing, so if you have that, you can include it.
(See sdcflows.utils.epimanip module - sdcflows 0+unknown documentation)
Note that EffectiveEchoSpacing accounts for ParallelReductionFactorInPlane. If you have the true echo spacing, you can include that as EchoSpacing along with ParallelReductionFactorInPlane and fmriprep will figure it out.
This stuff is fiendishly hard to get right manually. The simplest thing might be to rerun the protocol on a phantom, convert the DICOMs and use their metadata.
Hi @effigies,
Thanks so much, I get it now. I agree that manually entering the data is a hassle and can lead to mistakes. Your suggestions are really helpful, and I’ll give them a try right away!
Have a great weekend!
Best,
Shuning
Hi all,
I would like to ask if it’s possible to use the metadata files from other subjects for fMRIprep in the following situation: I have nifti format data for all 30 subjects, but I am missing the dicom format data for about 10 subjects. I want to convert the data to BIDS structure and use fMRIprep. Since the protocol parameters are the same for all subjects, can I use the metadata files from the subjects for whom I have dicom data for these 10 subjects?
Thank you!