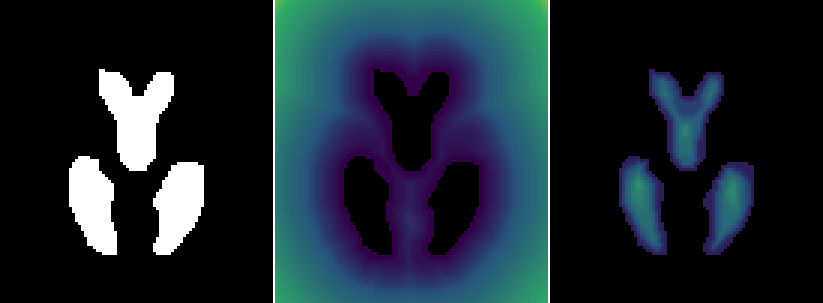Hello everyone,
I am using fsl bianca and LOcate for ms lesion segmentation.
I generated a binary ventricle mask from T1 image.
I need to create a distance map from the ventricles using FSL distancemap, it is too slow.
Can I use 'niimath vetnriclemask.niig.z -edt dxInVM’ for this aim? Is my command true?
fsl distance map output’s definition is = where the intensity of each voxel represent its distance in mm from the ventricles mask
Best
binary ventricle mask : ventricles:1 the remaining area:0
This topic is described here. The current FSL distancemap function uses an iterative algorithm that slows down exponentially with higher resolution images and also slows down exponentially s the number of dilations increases. In contrast, the niimath -edt Euclidean Transform uses a separable algorithm that scales linearly with resolution and where speed is not influenced by the size of the region. In the jiscmail example, for a typical size T1 MRI scan the difference was 36 hours for distancemap versus 1.5 seconds for niimath.
The good news is that future versions of FSL will adopt niimath’s algorithm, and it is also available in the new AFNI 3dDepthMap. Here is a Matlab function. For a description of the algorithm and Python code see Philip Rideout’s elegant page.
I already had a look at the thread you mentioned. I’m just confused here. regarding to your explanation… Which command is true to use?
niimath GM -edt dxInVM or niimath GM -binv -edt dxFromVM
the ventricles:1 the remaining area:0
Best, Dilek
The BIANCA documentation suggests distancemap <structural image>_vent.nii.gz dist_to_vent which would to niimath <structural image>_vent.nii.gz -binv -edt dist_to_vent. You can prove to yourself these generate identical results using the low resolution VentricleMask provided with FSL:
time distancemap -i $FSLDIR/data/standard/MNI152_T1_2mm_VentricleMask -o ~/dist_to_vent
time ./niimath $FSLDIR/data/standard/MNI152_T1_2mm_VentricleMask -binv -edt ~/ndist_to_vent
niimath ~/dist_to_vent --compare ~/ndist_to_vent; echo $?
While the results were identical, on my computer niimath was x2161 times faster and used 1/19th the RAM (with distancemap requiring 238 seconds and 194016 KB of memory).
If your masks are inverted, the command distancemap --invert -i <input> -o <output> can be translated as niimath <input> -edt <output>. Again, you can test that this generates identical results:
time distancemap --invert -i $FSLDIR/data/standard/MNI152_T1_2mm_VentricleMask -o ~/idist_to_vent
time niimath $FSLDIR/data/standard/MNI152_T1_2mm_VentricleMask -edt ~/nidist_to_vent
niimath ~/idist_to_vent --compare ~/nidist_to_vent; echo $?
For this example input image, distancemap is x1485 faster creating the invert than the standard image. This reveals that the iterative algorithm of distancemap is much slower if more iterations are required. Again, this sample image is intentionally very low resolution - since the distancemap algorithm gets exponentially slower with larger images, the performance difference between niimath and distancemap willl become more profound for real world datasets.
What do you mean while saying “If your masks are inverted”
Best, dilek
Given a binary image (left), distancemap provides the distance from the nearest bright voxel (middle) while distancemap --invert gives the distance from the nearest dark voxel (right). You can try it out yourself with the commands I provide above.
If you want further help with FSL tools, I suggest posting on the FSL jiscmail.
Continuing the discussion from Niimath usage for creating ventricle distancemap:
I exactly understood now.
I really appreciate for your time!
Best,
dilek