Hi Neuroimagers!
I am getting an affine error from nilearn when generating my second level model. The error is: ValueError: Field of view of image #31 is different from reference FOV.
A little bit about our data pipeline:
- I am working to process MID fMRI data from a particularly high motion sample.
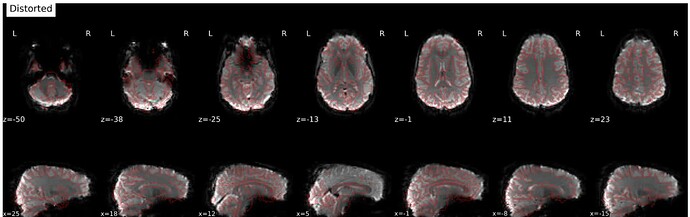
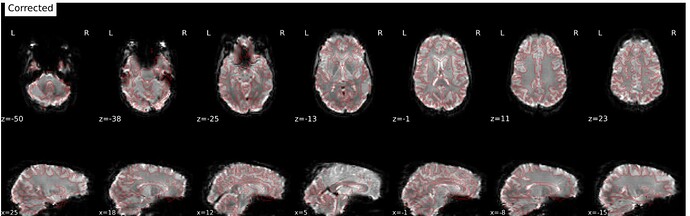
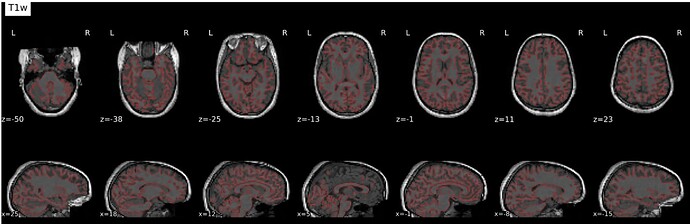
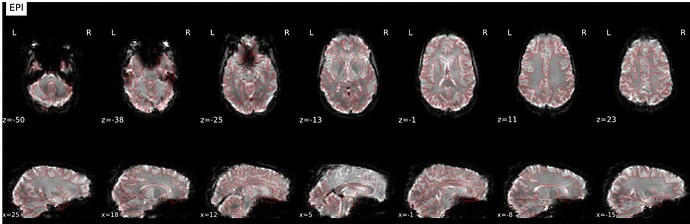
- Data is preprocessed using fmriprep 23.0.1, and all images are registered to MNI152NLin2009cAsym. The registration is decent, but we are having issues with areas of the preprocessed fmriprep output images showing brain outside of the standard MNI152NLin2009cAsym and it varies participant to participant.
- First level models and contrasts are created using
nilearn.FirstLevelModel. The mask image used in this FirstLevelModel call is the fmriprep participant and scan specific mask image. The mask images also show brain outside of the standard MNI space, and the outputted t-maps also show activation outside of the standard MNI space. - Finally, a second level model is run via
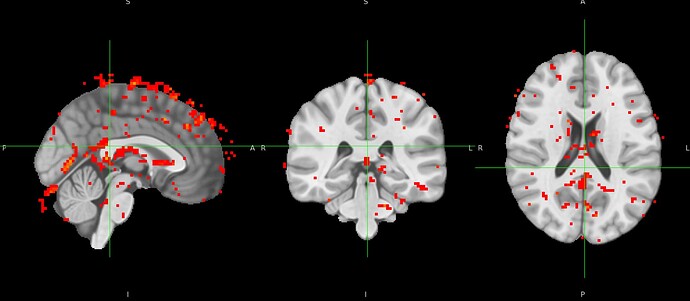
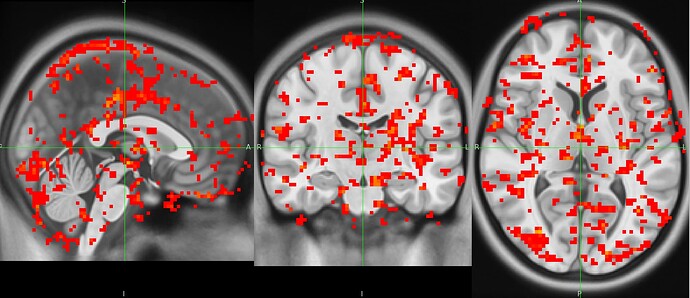
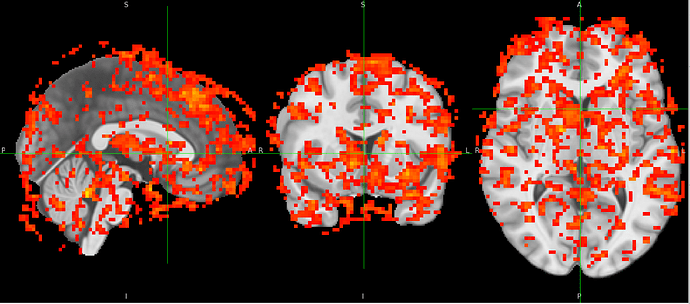
nilearn.SecondLevelModel, however errors out due to FOV error. When one with a smaller subsample, there is tons of activity shown outside of standard MNI space. I’ve pasted the affines below and included a screenshot.
Reference affine:
array([[ 2.5, 0. , 0. , -96.5],
[ 0. , 2.5, 0. , -132.5],
[ 0. , 0. , 2.5, -78.5],
[ 0. , 0. , 0. , 1. ]])
Image affine:
array([[ 2.5 , 0. , 0. , -96.5 ],
[ 0. , 2.5 , 0. , -132.5 ],
[ 0. , 0. , 2.50099993, -78.5 ],
[ 0. , 0. , 0. , 1. ]])
Reference shape:
(78, 93, 78)
Image shape:
(78, 93, 78, 1)
Below is a picture of a second level models run on a subsample that doesn’t include the participant with the problematic affines.
I read this thread but am wondering if resampling makes sense here and what image I would use, because in theory all images should already be in the same space. Is it possible that the fmriprep registering the MNI space isn’t super successful with these data? How big is too big of an affine difference for nilearn to tolerate?
Thanks for troubleshooting with me ![]()