Hi!
I used the Nilearn tools to visualize my data on a Jupyter Notebook.
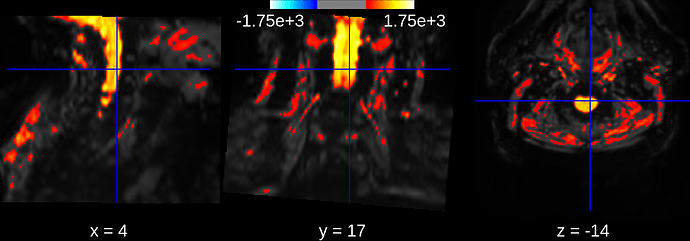
However, I have data from the spinal cord and when I display the spinal cord images on the background of a spinal cord anatomical image, I get a flattened image in the height direction for x and y slices, not the z slices. On the other hand, if I visualize this same image on an anatomical image including the brain and the spinal cord I don’t have this problem.
I used interactive plotting with plotting.view_img() with Nilearn 0.6.2
Do you have any suggestions to improve this visualization of this kind of data?
I try to modify some parameters in : ‘html_stat_map.py’ but I only success to modify the size of the viewer.
Thank for your help
Caroline
Hi, could you share a script and images to reproduce the problem? thanks!
I realize that sometimes it works well and sometimes not, but I don’t really understand why, for example with the 2 following functional mean images (3D):
When you plot the image1 under the image2, there is no problem.
But when you plot image2 under image1, the images are flattened
the code I use:
from nilearn import plotting
plotting.view_img(image2,bg_img=image1)
or
plotting.view_img(image1,bg_img=image2)
the images are here:
https://mcgill-my.sharepoint.com/:f:/g/personal/caroline_landelle_mcgill_ca/EqTA2DiUamVKh5jL2FkL0o0BkJ5weQCv-pm5wwmBSA279A?e=Tnb4Ye
tell me if you have some trouble to download them
Thanks
thanks! it seems that for some reason it doesn’t like the fact that image1 doesn’t have a diagonal affine. I think it is worth opening a bug report, but I suspect the issue may be in the (vendored) brainsprite javascript library rather than nilearn, as plot_stat_map seems to work fine with this image.
in the meanwhile you can solve the problem by resampling the image1 so it has a diagonal affine, e.g. image1 = nilearn.image.resample_to_img(image1, image2)
Thank you very much for your quick suggestions, but sometimes I have to vizualize two images, like image 1 (which do not have a diagonal affine). You can test for example by plotting image 1 under image 1 and this solution will not work.
I also try to resample individually the image1 using:
image.reorder_img(image1, resample=‘continuous’) as suggest by @GaelVaroquaux (https://github.com/nilearn/nilearn/issues/2546) but it didn’t work
sorry I’m not sure I understand why you cannot resample the image1? for example
import numpy as np
from nilearn import image, plotting
img1 = image.resample_img("image1.nii.gz", target_affine=np.eye(3))
plotting.view_img(img1, bg_img=img1, threshold="95%").open_in_browser()
Thanks Jerome,
image.reorder_img(image1, resample=‘continuous’)
didn’t solve the problem
But now it works if I resample the image as you suggest:
img1 = image.resample_img(“image1.nii.gz”, target_affine=np.eye(3))
glad to hear it! hopefully this bug will be fixed in a future nilearn release and then you won’t need this resampling anymore