Ahoi hoi @metaD,
you’re right that nilearn.decoding.searchlight is not happy with arrays, but expects (4D) Niimg-like objects. Hence, you would need to standardize your data before submitting it to the searchlight.
There are two ways (I think) of doing so: use NiftiMasker to standardize the data while converting it to a 2D array and then use inverse_transform to convert the standardized data back to a (4D) Niimg-like object OR use nilearn.image.clean_img to standardize your data keeping it in a (4D) Niimg-like object.
The respective functionality should become more clear within the following example code snippet based on the nilearn docs:
import pandas as pd
from nilearn import datasets
from nilearn.image import new_img_like, load_img, clean_img
from nilearn.input_data import NiftiMasker
# We fetch 2nd subject from haxby datasets (which is default)
haxby_dataset = datasets.fetch_haxby()
fmri_filename = haxby_dataset.func[0]
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
y = labels['labels']
session = labels['chunks']
# restrict example to certain conditions too shorten processing time
from nilearn.image import index_img
condition_mask = y.isin(['face', 'house'])
fmri_img = index_img(fmri_filename, condition_mask)
y, session = y[condition_mask], session[condition_mask]
# compute processing mask
import numpy as np
mask_img = load_img(haxby_dataset.mask)
# .astype() makes a copy.
process_mask = mask_img.get_data().astype(np.int)
picked_slice = 29
process_mask[..., (picked_slice + 1):] = 0
process_mask[..., :picked_slice] = 0
process_mask[:, 30:] = 0
process_mask_img = new_img_like(mask_img, process_mask)
# use niftimasker to standardize images
masker = NiftiMasker(standardize=True)
fmri_img_z = masker.fit_transform(fmri_img)
fmri_img_z = masker.inverse_transform(fmri_img_z)
# use clean_img to standardize images
fmri_img_clean_z = clean_img(fmri_img, standardize=True, detrend=False)
# setup searchlight for images standardized through niftimasker
import nilearn.decoding
searchlight = nilearn.decoding.SearchLight(
mask_img,
process_mask_img=process_mask_img,
radius=5.6, n_jobs=4,
verbose=1, cv=cv)
searchlight.fit(fmri_img_z, y)
# setup searchlight for images standardized through clean_img
searchlight_clean = nilearn.decoding.SearchLight(
mask_img,
process_mask_img=process_mask_img,
radius=5.6, n_jobs=4,
verbose=1, cv=cv)
searchlight_clean.fit(fmri_img_clean_z, y)
# compare both searchlight outcomes by visualizing them on the mean functional image
from nilearn import image
mean_fmri = image.mean_img(fmri_img)
from nilearn.plotting import plot_stat_map, plot_img, show
searchlight_img = new_img_like(mean_fmri, searchlight.scores_)
searchlight_img_clean = new_img_like(mean_fmri, searchlight_clean.scores_)
plot_img(searchlight_img, bg_img=mean_fmri,
title="Searchlight based on NiftiMasker", display_mode="z", cut_coords=[-9],
vmin=.42, cmap='hot', threshold=.2, black_bg=True)
plot_img(searchlight_img_clean, bg_img=mean_fmri,
title="Searchlight based on clean_img", display_mode="z", cut_coords=[-9],
vmin=.42, cmap='hot', threshold=.2, black_bg=True)
Here’s how the respective example outcomes look like:

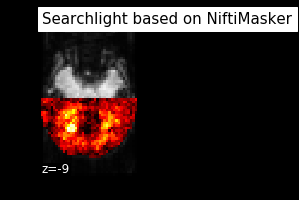
I would recommend clean_img for the sake of simplicity and time.
HTH, cheers, Peer

