I am using NiftiMasker to extract data from my images. I am interested only in gray matter. I have one control condition where I do not provide a mask image to mask_img. I set mask_strategy to mask_strategy='template', which according to nilearn documentations means, that it will compute a mask using nilearn.masking.compute_gray_matter_mask which will
“extract the gray matter part of your data by resampling the MNI152 brain mask for your data’s field of view”
Furthermore, in the documentation of nilearn.masking.compute_gray_matter_mask it says:
“Compute a mask corresponding to the gray matter part of the brain. The gray matter part is calculated through the resampling of MNI152 template gray matter mask onto the target image.”
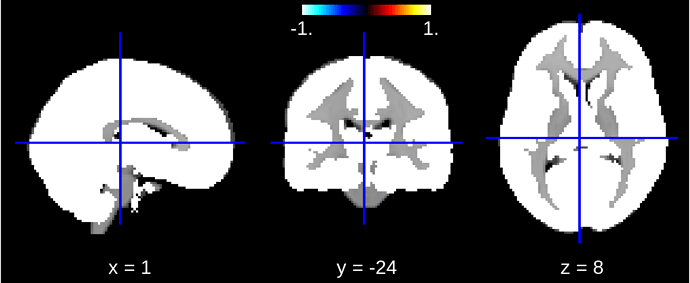
According to the documentation I expected the mask image to look something like this (this is a gray matter mask image provided from SDM):
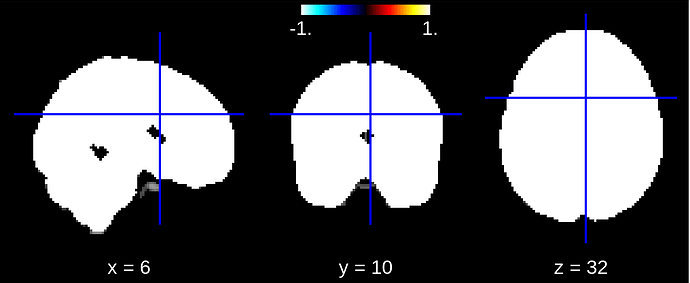
But when I look at the fitted mask_img_ in the NiftiMasker instance the mask looks like this:
Correct me if I am wrong, maybe I have a different conception about whole brain / gray matter and this is a misunderstanding, but the second mask image looks like a whole brain mask and not a gray matter mask. Can someone explain what is going on here?
Here’s my code:
from nilearn.datasets import fetch_oasis_vbm
from nilearn.input_data import NiftiMasker
from nilearn import plotting
# fetch OASIS dataset
oasis_img_paths = fetch_oasis_vbm(n_subjects=10)['gray_matter_maps']
# mask images and get data
niftimasker = NiftiMasker(mask_img=None,
smoothing_fwhm=8,
standardize=True,
mask_strategy='template',
memory='./niftimasker_cache')
X = niftimasker.fit_transform(oasis_img_paths)
# check used mask image
niftimasker_mask_img_ = niftimasker.mask_img_
html_view = plotting.view_img(niftimasker_mask_img_)
html_view.open_in_browser()