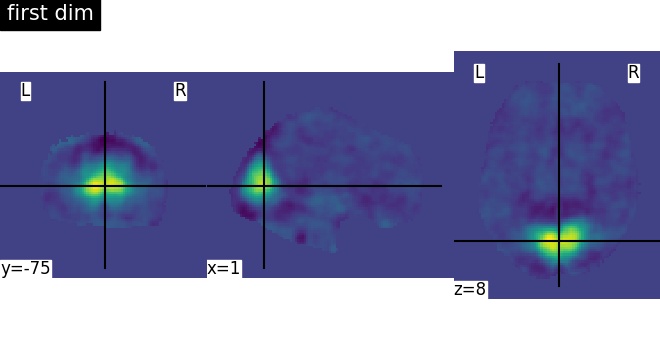
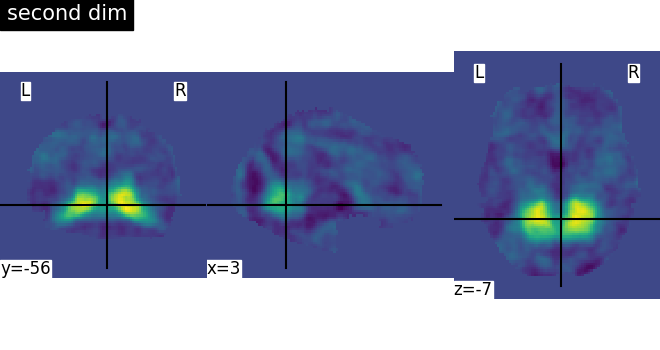
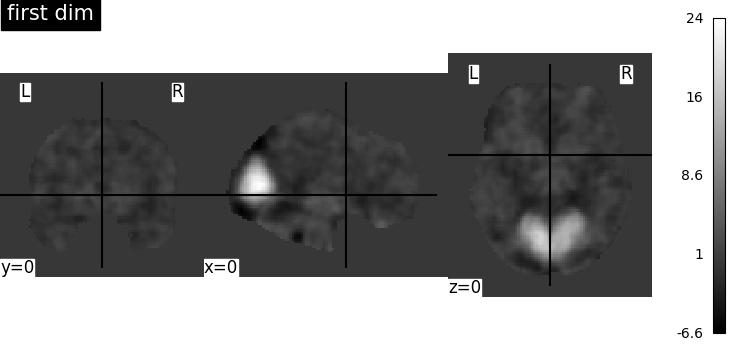
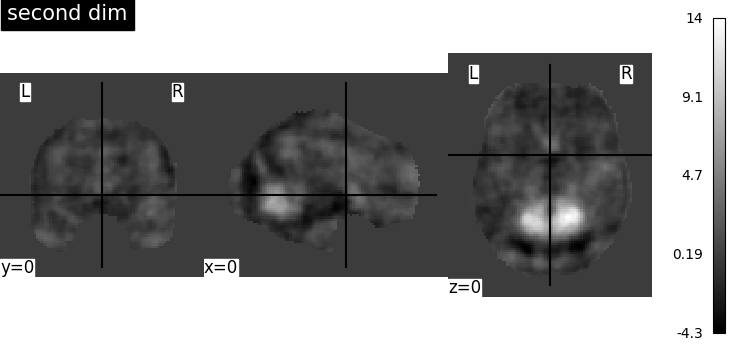
There seems to be an issue with the Smith network atlases imported with nilearn, each network image is the same. Is this a bug or am I missing something here?
Here’s a demonstration:
import nibabel as nb
import numpy as np
import pandas as pd
from nilearn import datasets
smith = datasets.fetch_atlas_smith_2009()
smith_70 = nb.load(smith["rsn70"])
smith70_dat = smith_70.get_fdata()
for i in range(69):
network_a = smith70_dat[:,:,:,i].astype(bool)
network_b = smith70_dat[:,:,:,i+1].astype(bool)
print(pd.Series((network_a == network_b).flatten()).value_counts())
import nilearn as nil
nil.__version__
Out[15]: '0.11.1'
The output comparing maps:
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64
True 902629
Name: count, dtype: int64