I’m currently working with using the LabelsMask and Connectome functionality in nilearn, and have been able to successfully run it on a new dataset with different parcellations (e.g. fetch_atlas_schaeffer2018, fetch_atlas_basc_multiscale_2015). However the results of the parcellation is that I get variable sized tensors. For example, if I’m using the 64 segment parcellation, I may get results with 62,63, or sometimes even 60.
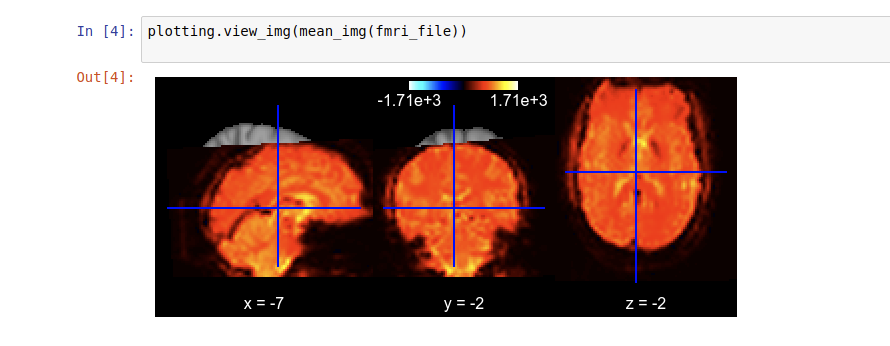
My understanding is that this might be due to the data may not aligned properly in the space, which can always be a problem presented when using new data. I used the plotting function of nilearn overlayed with the mean image of the fmri data, and it does appear to not be aligned.
What would be the easiest way to align the data into the same space during thepreprocessing step? Wonder if I need to perform registration on the data.