Hi everyone,
I’ve been stuck on my Level1Design node creating incorrect ev_files. Specifically, instead of generating 11 EV files, it generates 7: 1 for the first ev and then rest are txt files combined with the Outliers and realignment_parameters txt files that are input into SpecifyModel. When I run a single participant, node by node, on jupyter notebook, the correct ev_files are created. However, it just doesn’t seem to work on my terminal. I’m probably missing something super obvious and could use a fresh pair of eyes.
Thanks in advance!
CODE:
Level 1 TMS BLOCK Designs
###########################################
Purpose: Create TMS on and TMS off Model
for Individual then Normalize data
for group effects
METHOD:
1. Get dependencies
2. Set Experiment Parameters
1. Import Dependencies
#%matplotlib inline
from os.path import join as opj
import os.path
import json
from nipype.interfaces.fsl import (Level1Design, FEATModel, L2Model,
FEAT, ContrastMgr, FILMGLS, Merge, GLM, FEATRegister, Randomise, FLAMEO)
#from nipype.algorithms.modelgen import SpecifySPMModel
from nipype.interfaces.utility import Function, IdentityInterface
from nipype.interfaces.io import SelectFiles, DataSink
from nipype.pipeline.engine import Workflow, Node, MapNode
from nipype.algorithms.modelgen import SpecifyModel
import pandas as pd
from nipype.interfaces.base import Bunch
2.Set Experiment Specific Attributes
preproc_dir = ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2’
output_dir = ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/LEVELONE’ #need to make this
working_dir = ‘workingdir_LEVELONE’
#Could change this to be the actual name of the folders
subject_list= [‘P100’,‘P101’]
‘P101’, ‘P102’, ‘P103’, ‘P104’, ‘P105’,
‘P106’, ‘P107’, ‘P108’, ‘P109’, ‘P110’, ‘P111’,
‘P201’, ‘P202’, ‘P203’, ‘P204’, ‘P205’, ‘P206’,
‘P207’, ‘P208’, ‘P209’, ‘P210’, ‘P211’, ‘P212’,
‘P213’, ‘P214’, ‘P215’, ‘P216’]
ROI_list = [‘L_DLPFC_BLOCK_TMS’, ‘R_DLPFC_BLOCK_TMS’]
TR = 2.5
hpcutoff =100
3. ModelPrep Workflow
modelprep = Workflow(name = ‘modelprep’)
# Specify Model#
# INPUTS NEEDED: event_files, functional_runs
# high_pass_filter_cutoff, input_units: (u'secs' or u'scans')
# subject_info (from getsubject info), time_repetition (TR = 2.5)
# Need to add outlier file as part of workflow from selectFiles
modelspec = Node(SpecifyModel(high_pass_filter_cutoff= hpcutoff,
input_units = ‘secs’, time_repetition = TR),
name=“modelspec”) #multiple EV files so MapNode?
#1)-Get subject specific BEHAVE Files (In this case its the same for everyone)-#
# Generate Design Matrix
# INPUTS NEEDED: Interscan interval (TR), bases (gamma, hrf)
#session info (from Specify Model)
level1design = Node(Level1Design(interscan_interval = TR, bases = {‘dgamma’: {‘derivs’: False}},
model_serial_correlations = True),name = “level1design”)
#contrasts can be gotten from infosource
# Generate MAT files
#INPUT NEEDED: ev_files and fsf_file from LEVEL1Design
modelgen = Node(FEATModel(),name=‘modelgen’,
#iterfield=[‘fsf_file’, ‘ev_files’]
)
# Generate Contrast Files
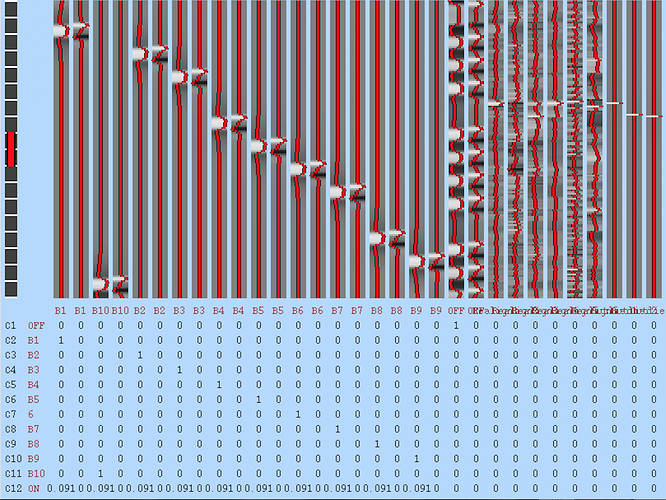
condition_names = [‘OFF’, ‘B1’, ‘B2’, ‘B3’, ‘B4’, ‘B5’, ‘B6’,‘B7’,‘B8’,‘B9’,‘B10’]
Contrast01= [‘OFF’, ‘T’, condition_names, [1,0,0,0,0,0,0,0,0,0,0]]
Contrast02= [‘Block1’,‘T’, condition_names, [0,1,0,0,0,0,0,0,0,0,0]]
Contrast03= [‘Block2’,‘T’, condition_names, [0,0,1,0,0,0,0,0,0,0,0]]
Contrast04= [‘Block3’,‘T’, condition_names, [0,0,0,1,0,0,0,0,0,0,0]]
Contrast05= [‘Block4’,‘T’, condition_names, [0,0,0,0,1,0,0,0,0,0,0]]
Contrast06= [‘Block5’,‘T’, condition_names, [0,0,0,0,0,1,0,0,0,0,0]]
Contrast07= [‘Block6’,‘T’, condition_names, [0,0,0,0,0,0,1,0,0,0,0]]
Contrast08= [‘Block7’,‘T’, condition_names, [0,0,0,0,0,0,0,1,0,0,0]]
Contrast09= [‘Block8’,‘T’, condition_names, [0,0,0,0,0,0,0,0,1,0,0]]
Contrast10= [‘Block9’,‘T’, condition_names, [0,0,0,0,0,0,0,0,0,1,0]]
Contrast11= [‘Block10’,‘T’, condition_names,[0,0,0,0,0,0,0,0,0,0,1]]
Contrast12= [‘ON’, ‘T’, condition_names,[0,1/11.,1/11.,1/11.,1/11.,1/11.,1/11.,1/11.,1/11.,1/11.,1/11.]]
contrast_list= [Contrast01,Contrast02,Contrast03,Contrast04,Contrast05,Contrast06,
Contrast07,Contrast08,Contrast09,Contrast10,Contrast11]
# Connect ModelPrep WorkLoad
modelprep.base_dir=preproc_dir
modelprep.connect([(modelspec, level1design, [(‘session_info’, ‘session_info’)]),
(level1design, modelgen, [(‘fsf_files’, ‘fsf_file’),(‘ev_files’, ‘ev_files’)]),
])
4 Model FIT workgflow (where stats gets done!)
#modelfit = Workflow(name = ‘modelfit’)
Generate model estimates based on fsf and mat files from above
# in_file fomes from SelectFiles (its the functional run)
#design file from Level1Design or FEATModel
modelestimate = MapNode(FILMGLS(smooth_autocorr=True, mask_size=5, threshold=1000),
name=‘modelestimate’,
iterfield=[‘design_file’, ‘in_file’, ‘tcon_file’])
# Generate contrast estimates DONT NEED THIS ANYMORE FILM DOES COPES
conestimate = MapNode(ContrastMgr(),name=‘conestimate’,
iterfield=[
‘tcon_file’, ‘param_estimates’, ‘sigmasquareds’,
‘dof_file’
])
# Link modefit workflow for Level 1
modelfit.connect([
(modelestimate, conestimate, [(‘param_estimates’, ‘param_estimates’),
(‘sigmasquareds’,‘sigmasquareds’),(‘dof_file’, ‘dof_file’)]),
])
5 Input and outputs
infosource = Node(IdentityInterface(fields=[‘subject_id’,
‘ROI_id’, ‘contrasts’], contrasts=contrast_list),
name=“infosource”)
infosource.iterables = [(‘subject_id’, subject_list),
(‘ROI_id’, ROI_list),]
##THIS NEEDS TO BE CHANGED TO PROPER NAMES####
anat_file = opj(‘preproc’,’{ROI_id}{subject_id}’, ‘Anat_Orient_brain.nii.gz’)
func_file = opj(‘preproc’,’{ROI_id}{subject_id}’, ‘Func_Ready_mcf_st_flirt_smooth.nii.gz’)
art_file=opj(‘preproc’,’{ROI_id}{subject_id}’,‘art.Func_Ready_mcf_st_flirt_outliers.txt’)
motionCorrection_parameters=opj(‘preproc’,’{ROI_id}{subject_id}’,‘Func_Ready_mcf.nii.par’)
templates = {‘anat’: anat_file,
‘func’: func_file,
‘art’: art_file,
‘mc_par’: motionCorrection_parameters}
selectfiles = Node(SelectFiles(templates,
base_directory=preproc_dir,
sort_filelist=True),
name=“selectfiles”)
# Get information for workflows
def subjectinfo(subject_id):
import pandas as pd
from nipype.interfaces.base import Bunch
from os.path import join as opj
#EV_FILE= opj(‘preproc’,‘TMS_Block_Timings.txt’)
trialinfo = pd.read_table(’~/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/preproc/TMS_Block_Timings.txt’)
trialinfo.head()
conditions = []
onsets = []
durations = []
for group in trialinfo.groupby(‘Event’):
conditions.append(group[0])
onsets.append(group[1].Onset.tolist())
durations.append(group[1].Duration.tolist())
subject_info = [Bunch(conditions=conditions,
onsets=onsets,durations=durations
#amplitudes=None,
#tmod=None,
#pmod=None,
#regressor_names=None,
#regressors=None
)]
return subject_info # this output will later be returned to infosource
# Function node to use getsubjectinfo
getsubjectinfo = Node(Function(input_names=[‘subject_id’],
output_names=[‘subject_info’],
function=subjectinfo),
name=‘getsubjectinfo’)
# Set outputs
datasink = Node(DataSink(base_directory=preproc_dir,
container=output_dir),
name=“datasink”)
substitutions = [(’_subject_id’, ‘’),
(‘ROI_id’, ‘’)]
datasink.inputs.substitutions = substitutions
#Figure out substitutions for functional and anatomicals
6 Level1Pipeline Workflow
LEVELONE = Workflow(name=‘LEVELONE’)
LEVELONE.base_dir= preproc_dir
LEVELONE.connect([
(infosource, selectfiles, [(‘subject_id’, ‘subject_id’),
(‘ROI_id’, ‘ROI_id’), (‘contrasts’, ‘contrasts’)]),
(infosource, getsubjectinfo, [(‘subject_id’, ‘subject_id’)]),
(getsubjectinfo, modelprep, [(‘subject_info’, ‘modelspec.subject_info’)]),
(selectfiles, modelprep, [(‘func’, ‘modelspec.functional_runs’)]),
(selectfiles, modelprep, [(‘art’,‘modelspec.outlier_files’),
(‘mc_par’, ‘modelspec.realignment_parameters’)]),
(infosource, modelprep, [(‘contrasts’, ‘level1design.contrasts’)]),#inputs contasts into level1design
(selectfiles, modelestimate, [(‘func’, ‘in_file’)]),
(modelprep, modelestimate, [(‘modelgen.design_file’, ‘design_file’)]),
(modelprep, modelestimate, [(‘modelgen.con_file’, ‘tcon_file’)]),
# (selectfiles, modelfit, [('func', 'modelestimate.in_file')]),
# (modelprep, modelfit, [('modelgen.design_file', 'modelestimate.design_file')]),# could put these two lines in pipelien alter
# (modelprep, modelfit, [('modelgen.con_file', 'conestimate.tcon_file')]),
(modelprep, datasink, [('level1design.fsf_files','LEVELONE.@fsf_files'),
('level1design.ev_files', 'LEVELONE.@ev_files')]),
(modelestimate, datasink, [('logfile', 'LEVELONE.@logfile'),
('param_estimates', 'LEVELONE.@param_estimates'),
('residual4d', 'LEVELONE.@residual4d'),
('thresholdac', 'LEVELONE.@thresholdac'),
('copes', 'LEVELONE.@copes'),
('varcopes', 'LEVELONE.@varcopes'),
('zstats', 'LEVELONE.@zstats'),
('tstats', 'LEVELONE.@tstats')]),
# (modelestimate, datasink, [('copes', 'LEVELONE.@copes'),
# ('varcopes', 'LEVELONE.@varcopes'),
# ('zstats', 'LEVELONE.@zstats'),
# ('tstats', 'LEVELONE.@tstats')]),
])
Generate Graph and execute
LEVELONE.write_graph()
LEVELONE.run()
Crashfile:
iMac:fMRI_data marlengonzalez$ nipypecli crash crash-20180407-170629-marlengonzalez-modelgen.a3-8475f95e-b60b-4ed8-a9df-a418f4bafcea.pklz
File: /Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/crash-20180407-170629-marlengonzalez-modelgen.a3-8475f95e-b60b-4ed8-a9df-a418f4bafcea.pklz
Node: LEVELONE.modelprep.modelgen
Working directory: /Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen
Node inputs:
args =
environ = {‘FSLOUTPUTTYPE’: ‘NIFTI_GZ’}
WHERE EVERYTHING GOES WRONG. WHAT ARE THESE???]###
ev_files = [’/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_B1_0_1.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign1_0_2.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign2_0_3.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign3_0_4.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign4_0_5.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign5_0_6.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Realign6_0_7.txt’, ‘/Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/ev_Outlier1_0_8.txt’]
fsf_file = /Users/marlengonzalez/Documents/Projects/spTMS-fMRI/fMRI_data/PREPROC_Try2/LEVELONE/modelprep/_ROI_id_R_DLPFC_BLOCK_TMS_subject_id_P101/modelgen/run0.fsf
ignore_exception = False
output_type = NIFTI_GZ
terminal_output =
Traceback:
Traceback (most recent call last):
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/pipeline/plugins/linear.py”, line 44, in run
node.run(updatehash=updatehash)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 487, in run
result = self._run_interface(execute=True)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 571, in _run_interface
return self._run_command(execute)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/pipeline/engine/nodes.py”, line 650, in _run_command
result = self._interface.run(cwd=outdir)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/interfaces/base/core.py”, line 516, in run
runtime = self._run_interface(runtime)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/interfaces/base/core.py”, line 1023, in _run_interface
self.raise_exception(runtime)
File “/Users/marlengonzalez/anaconda3/lib/python3.6/site-packages/nipype/interfaces/base/core.py”, line 960, in raise_exception
).format(**runtime.dictcopy()))
RuntimeError: Command:
feat_model run0
Standard output:
Standard error:
Contrast 1 is empty!
Return code: 1