Hello everyone,
I am trying to run bedpostx_parallel workflow in nipype. It works fine during my test runs on downsampled data, however when I try to run it on the original data it fails at the step where it tries to merge all the outputs from parallel nodes into single files.
I looked into the issue, and it seems that on this line
the output of np.squeeze is a 0D array, and getting the length of 0D array isn’t possible in
I am wondering whether this is just an anomaly due to the heuristic nature of the algorithm and would probably not be an issue on a subsequent run or is it something that needs to be handled within nipype, for example, via:
np.atleast_1d
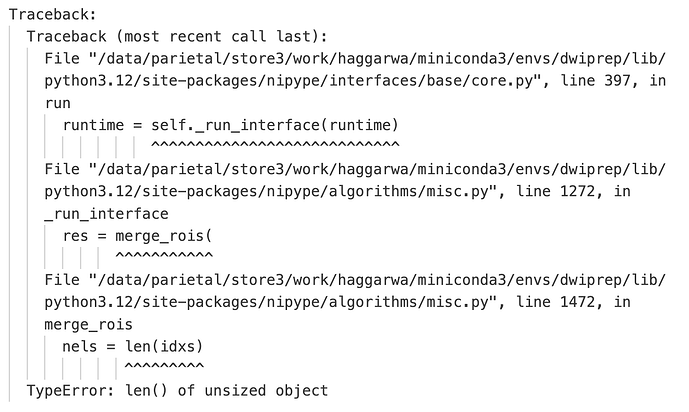
Here’s the traceback of log from the crash file. I am running python 3.12 and nipype 1.8.6.
Please let me know if you need more info.
Thanks!