Hello, Nipype experts:
Recently, I want to use TRACULA pipeline in nipype, and luckily I found that Satra has already done this, this is the link: https://gist.github.com/satra/82dbf385bcba9d97fd9b
So I adjust his script to use for my data, but I always got some errors to say that:
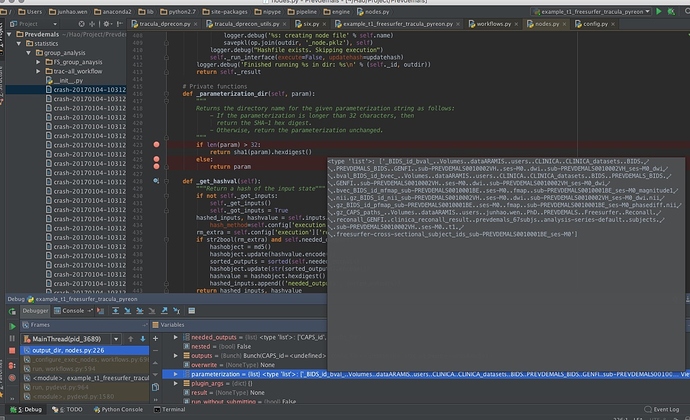
OSError: Could not create /Users/junhao.wen/test/test-tracula/trac-all_workflow/_BIDS_id_bval_..Volumes..dataARAMIS..users..CLINICA..CLINICA_datasets..BIDS..PREVDEMALS_BIDS..GENFI..sub-PREVDEMALS0010001BE..ses-M0..dwi..sub-PREVDEMALS0010001BE_ses-M0_dwi.bval_BIDS_id_bvec_..Volumes..dataARAMIS..users..CLINICA..CLINICA_datasets..BIDS..PREVDEMALS_BIDS..GENFI..sub-PREVDEMALS0010001BE..ses-M0..dwi..sub-PREVDEMALS0010001BE_ses-M0_dwi.bvec_BIDS_id_mfmap_sub-PREVDEMALS0010001BE..ses-M0..fmap..sub-PREVDEMALS0010001BE_ses-M0_magnitude1.nii.gz_BIDS_id_nii_sub-PREVDEMALS0010001BE..ses-M0..dwi..sub-PREVDEMALS0010001BE_ses-M0_dwi.nii.gz_BIDS_id_pfmap_sub-PREVDEMALS0010002VH..ses-M0..fmap..sub-PREVDEMALS0010002VH_ses-M0_phasediff.nii.gz_CAPS_paths_..Volumes..dataARAMIS..users..junhao.wen..PhD..PREVDEMALS..Freesurfer..Reconall..reconall_GENFI..clinica_reconall_result..prevdemals_67subjs..analysis-series-default..subjects..sub-PREVDEMALS0010001BE..ses-M0..t1..freesurfer-cross-sectional_subject_ids_sub-PREVDEMALS0010002VH_ses-M0/trac-prep
So I dig into the code, I found the problem is from the private function from nipype.Node._parameterization_dir(param) :
Private functions
def _parameterization_dir(self, param): """ Returns the directory name for the given parameterization string as follows: - If the parameterization is longer than 32 characters, then return the SHA-1 hex digest. - Otherwise, return the parameterization unchanged. """ if len(param) > 32: return sha1(param).hexdigest() else: return param
It seems that in the nipype.Function interface, when we define the working_directory, the node.parameterization is too long, so that it can not create the folder. And the path is in a mess… In Satra’s script, he use nipype.Function to call a function, for example run_prep, and in the run_prep, he created another node to call the command line for TRACULA.
Before, I tried to wrap another interface, it works well, but this time, when I debug this script, here is the output of sub-task of my workflow for the paramerization func:
> trac_all_worlflow.trac-prep.a009.parameterization=
> <type 'list'>: ['_BIDS_id_bval_..Volumes..dataARAMIS..users..CLINICA..CLINICA_datasets..BIDS..PREVDEMALS_BIDS..GENFI..sub-PREVDEMALS0010001BE..ses-M0..dwi..sub-PREVDEMALS0010001BE_ses-M0_dwi.bval_BIDS_id_bvec_..Volumes..dataARAMIS..users..CLINICA..CLINICA_datasets..BIDS..PREVDEMALS_BIDS..GENFI..sub-PREVDEMALS0010002VH..ses-M0..dwi..sub-PREVDEMALS0010002VH_ses-M0_dwi.bvec_BIDS_id_mfmap_sub-PREVDEMALS0010001BE..ses-M0..fmap..sub-PREVDEMALS0010001BE_ses-M0_magnitude1.nii.gz_BIDS_id_nii_sub-PREVDEMALS0010001BE..ses-M0..dwi..sub-PREVDEMALS0010001BE_ses-M0_dwi.nii.gz_BIDS_id_pfmap_sub-PREVDEMALS0010002VH..ses-M0..fmap..sub-PREVDEMALS0010002VH_ses-M0_phasediff.nii.gz_CAPS_paths_..Volumes..dataARAMIS..users..junhao.wen..PhD..PREVDEMALS..Freesurfer..Reconall..reconall_GENFI..clinica_reconall_result..prevdemals_67subjs..analysis-series-default..subjects..sub-PREVDEMALS0010001BE..ses-M0..t1..freesurfer-cross-sectional_subject_ids_sub-PREVDEMALS0010001BE_ses-M0']
Any suggestions will be appreciated:)
Happy christmas
Hao