Dear experts,
@e.urunuela @dowdlelt
I am seeking assistance with a persistent error occurring in a subset of subjects during our task-based fMRI preprocessing pipeline using AFNI’s afni_proc.py and multi-echo TE-dependence analysis (tedana).
The Problem
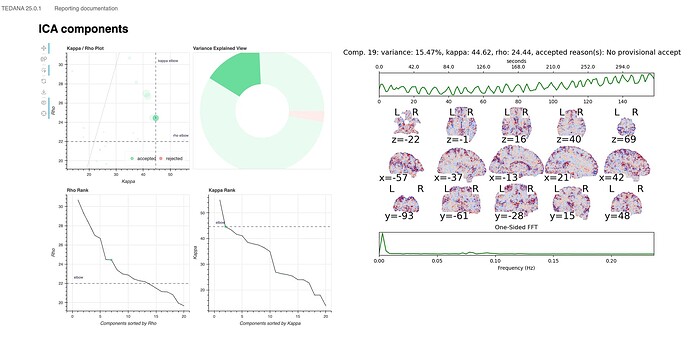
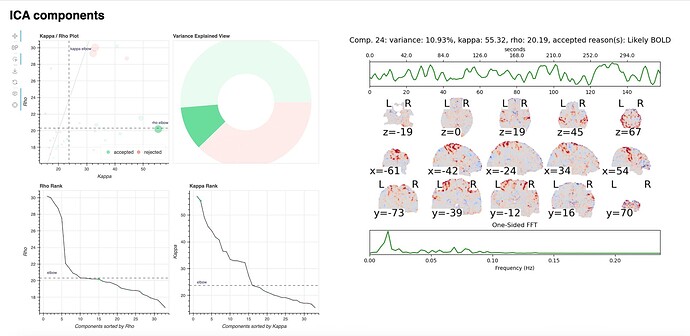
For a number of subjects, the tedana component returns the critical warning/error: “No BOLD components detected!”
- Other subjects run successfully without this issue.
- I have verified that the subjects with the error do not exhibit unusually high motion compared to the successful subjects.
Acquisition Details
- Field Strength: 7 Tesla
- Voxel Size: 1.5 mm isotropic
- Analysis: Task-based fMRI
Processing Details
I am calling tedana via afni_proc.py using the following option:
-combine_method m_tedana
The resulting generated tedana command, which is failing for these subjects, is:
tedana -d ${d1} ${d2} ${d3} -e 14.0 37.87 61.74 --mask ${mask_epi_anat} --maxit 5000 --tedpca kundu-stabilize --fittype curvefit --tree meica --n-threads 30 --ica_method robustica --n_robust_runs 30 --maxrestart 500 --lowmem --gscontrol mir gsr --out-dir tedana_r01 --convention orig
Troubleshooting Attempted
I have tried the following variations, but the error persists for the specific subset of subjects:
- Changing --tedpca options to “AIC” and “KIC”.
- Changing --tree to “tedana_orig”.
Example Data
I uploaded an example run (and BIDS .json for the echo1 dataset)that exhibits this error here: https://www.dropbox.com/scl/fo/439bttcbln35ywk9xlnqo/AMIVcj6P4OIFi_uRhfndCEM?rlkey=m0ti3pqf7t1ws0nssq9kw83fr&st=vvsj7q69&dl=0
Here is the afni_proc.py command:
afni_proc.py -subj_id 816 -scr_overwrite
-dsets_me_run ${run1echo1} ${run1echo2} ${run1echo3}
-dsets_me_run ${run2echo1} ${run2echo2} ${run2echo3}
-dsets_me_run ${run3echo1} ${run3echo2} ${run3echo3} -dsets_me_run ${run4echo1} ${run4echo2} ${run4echo3}
-echo_times 14.0 37.87 61.74 -out_dir ${outdir}
-blocks despike tshift align tlrc volreg mask combine blur scale
regress -blip_forward_dset ${AP}
-blip_reverse_dset ${PA} -blur_size 3
-blur_in_mask yes
-copy_anat T1_bc_SS.nii.gz -anat_has_skull no -mask_opts_automask -clfrac 0.3 -dilate 1
-mask_epi_anat yes
-tshift_opts_ts -tpattern @SliceTiming.1D -align_unifize_epi local
-align_opts_aea -cost lpc+ZZ -giant_move -check_flip
-epi_strip 3dAutomask
-tlrc_base MNI152_2009_template_SSW.nii.gz \
-tlrc_NL_warp \
-tlrc_NL_warped_dsets anatQQ.816.nii \ anatQQ.816.aff12.1D \
anatQQ.816_WARP.nii \
-volreg_align_to MIN_OUTLIER -volreg_align_e2a -volreg_tlrc_warp \ -regress_apply_mot_types demean deriv -regress_motion_per_run \ -regress_censor_motion 0.3 -regress_censor_outliers 0.15 \
-regress_stim_times
{regress options}
-combine_method m_tedana
-combine_tedana_path /hpc/users/chowda26/.local/bin/tedana
-combine_opts_tedwrap -tedana_is_exec
-combine_opts_tedana '--maxit 5000 --tedpca kundu-stabilize
--fittype curvefit --tree meica --n-threads 30
--ica_method robustica --n_robust_runs 30
--maxrestart 500 --lowmem --gscontrol {mir,gsr}'
Any insight into why tedana might be failing to detect BOLD components only in this subset of 7T data would be greatly appreciated.
Thank you,
Avi