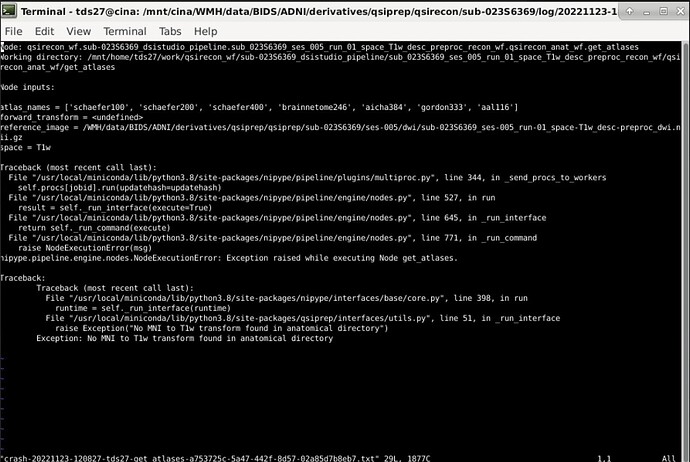
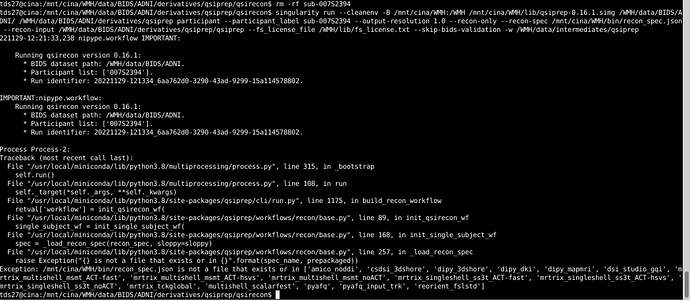
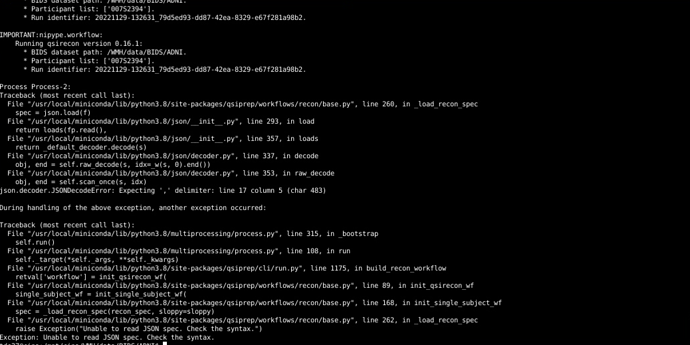
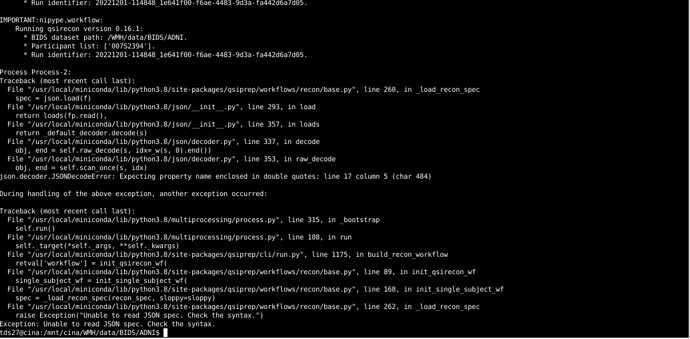
For your QSIRecon command, I would set the output dir and recon-input as /WMH/data/BIDS/ADNI/derivatives/, unless the qsiprep outputs are in a folder called derivatives/qsiprep/qsiprep
Also, lets try only having one recon-spec. You can combine the amico_noddi and dsi_studio_gqi workflows into one recon_spec by combining their jsons, like so:
{ "description": "Runs the AMICO implementation of NODDI",
"space": "T1w",
"name": "amico_noddi",
"atlases": ["schaefer100", "schaefer200", "schaefer400", "brainnetome246", "aicha384", "gordon333", "aal116"],
"nodes": [
{
"name": "fit_noddi",
"action": "fit_noddi",
"software": "AMICO",
"input": "qsiprep",
"output_suffix": "NODDI",
"parameters": {
"isExvivo": false,
"dPar": 1.7E-3,
"dIso": 3.0E-3
}
{
"name": "dsistudio_gqi",
"software": "DSI Studio",
"action": "reconstruction",
"input": "qsiprep",
"output_suffix": "gqi",
"parameters": {"method": "gqi"}
},
{
"name": "scalar_export",
"software": "DSI Studio",
"action": "export",
"input": "dsistudio_gqi",
"output_suffix": "gqiscalar"
},
{
"name": "tractography",
"software": "DSI Studio",
"action": "tractography",
"input": "dsistudio_gqi",
"parameters": {
"turning_angle": 35,
"method": 0,
"smoothing": 0.0,
"step_size": 1.0,
"min_length": 30,
"max_length": 250,
"seed_plan": 0,
"interpolation": 0,
"initial_dir": 2,
"fiber_count": 5000000
}
},
{
"name": "streamline_connectivity",
"software": "DSI Studio",
"action": "connectivity",
"input": "tractography",
"output_suffix": "gqinetwork",
"parameters": {
"connectivity_value": "count,ncount,mean_length,gfa",
"connectivity_type": "pass,end"
}
}
]
}
You can save that as a .json file, and put the path to that as your --recon-spec, and make sure it is contained in a path that is bound to the container (somewhere in /mnt/cina/WMH/).
So, after checking to make sure QSIPrep ran successfully for these subjects (e.g., checking the HTML outputs, I would run this command for QSIRecon, using a fresh work directory:
singularity run --cleanenv -B /mnt/cina/WMH:/WMH /mnt/cina/WMH/lib/qsiprep-0.16.1.simg /WMH/data/BIDS/ADNI/ /WMH/data/BIDS/ADNI/derivatives/ participant --participant_label sub-036S6179 --recon-only --recon-spec /PATH/TO/YOUR/RECON/SPEC.json --recon-input /WMH/data/BIDS/ADNI/derivatives/qsiprep/ --fs_license_file /WMH/lib/fs_license.txt --skip-bids-validation -w SPECIFY/WORK/DIR