Summary of what happened:
Hi,
I’m trying to use fitlins for the first try and I’m struggling with building the model in the JSON file. I have a task that has been measured over 3 sessions (out of 6) for each participant in two different groups. I tried to start easy and build a model for 2 participants just with the contrast (no movement regressors, whatsoever) but it seems I’m still doing something wrong in the JSON file and I can’t figure it out.
Command used (and if a helper script was used, a link to the helper script or the command generated):
fitlins /dataset/rawdata /dataset/analyzed participant \
-d /dataset/derivatives/fmriprep -w fitlins_cache \
--smoothing 6:dataset:iso -m /dataset/rawdata/models/model-faces_smdl.json
Version:
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
bids compatible, my derivatives folder comes from fmriPrep
Relevant log outputs (up to 20 lines):
The error I get when running fitlins:
fitlins /dataset/rawdata /dataset/analyzed participant -d /dataset/derivatives/fmriprep -w fitlins_cache --smoothing 6:dataset:iso -m /dataset/rawdata/models/model-faces_smdl.json
Captured warning (<class 'UserWarning'>): `--estimator nistats` is a deprecated synonym for `--estimator nilearn`. Future versions will raise an error.
Captured warning (<class 'UserWarning'>): The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
240315-12:15:32,595 nipype.workflow INFO:
[Node] Setting-up "fitlins_wf.loader" in "/zi/home/miroslava.jindrova/fitlins_cache/fitlins_wf/loader".
240315-12:15:32,623 nipype.workflow INFO:
[Node] Executing "loader" <fitlins.interfaces.bids.LoadBIDSModel>
/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/layout/validation.py:156: UserWarning: The PipelineDescription field was superseded by GeneratedBy in BIDS 1.4.0. You can use ``pybids upgrade`` to update your derivative dataset.
warnings.warn("The PipelineDescription field was superseded "
240315-12:17:03,654 nipype.workflow INFO:
[Node] Finished "loader", elapsed time 91.015491s.
240315-12:17:03,655 nipype.workflow WARNING:
Storing result file without outputs
240315-12:17:03,662 nipype.workflow WARNING:
[Node] Error on "fitlins_wf.loader" (/zi/home/miroslava.jindrova/fitlins_cache/fitlins_wf/loader)
240315-12:17:04,267 nipype.workflow ERROR:
Node loader failed to run on host zislrds0068.zi.local.
240315-12:17:04,268 nipype.workflow ERROR:
Saving crash info to /zi/home/miroslava.jindrova/fitlins_cache/crash-20240315-121704-miroslava.jindrova-loader-43f6b8c7-b1f6-463d-af78-42c0a68d3551.txt
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node loader.
Traceback:
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 248, in _run_interface
self._results['all_specs'] = self._load_graph(runtime, graph)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 256, in _load_graph
specs = node.run(inputs, group_by=node.group_by, **filters)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 471, in run
node_output = BIDSStatsModelsNodeOutput(
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 593, in __init__
df = reduce(pd.DataFrame.merge, dfs)
TypeError: reduce() of empty sequence with no initial value
240315-12:17:06,266 nipype.workflow ERROR:
could not run node: fitlins_wf.loader
FitLins failed: Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node loader.
Traceback:
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 248, in _run_interface
self._results['all_specs'] = self._load_graph(runtime, graph)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 256, in _load_graph
specs = node.run(inputs, group_by=node.group_by, **filters)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 471, in run
node_output = BIDSStatsModelsNodeOutput(
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 593, in __init__
df = reduce(pd.DataFrame.merge, dfs)
TypeError: reduce() of empty sequence with no initial value
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/bin/fitlins", line 8, in <module>
sys.exit(main())
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/cli/run.py", line 442, in main
sys.exit(run_fitlins(sys.argv[1:]))
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/cli/run.py", line 419, in run_fitlins
fitlins_wf.run(**plugin_settings)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/plugins/base.py", line 212, in run
raise error from cause
RuntimeError: Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node loader.
Traceback:
Traceback (most recent call last):
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/nipype/interfaces/base/core.py", line 398, in run
runtime = self._run_interface(runtime)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 248, in _run_interface
self._results['all_specs'] = self._load_graph(runtime, graph)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/fitlins/interfaces/bids.py", line 256, in _load_graph
specs = node.run(inputs, group_by=node.group_by, **filters)
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 471, in run
node_output = BIDSStatsModelsNodeOutput(
File "/opt/miniconda-latest/envs/neuro/lib/python3.9/site-packages/bids/modeling/statsmodels.py", line 593, in __init__
df = reduce(pd.DataFrame.merge, dfs)
TypeError: reduce() of empty sequence with no initial value
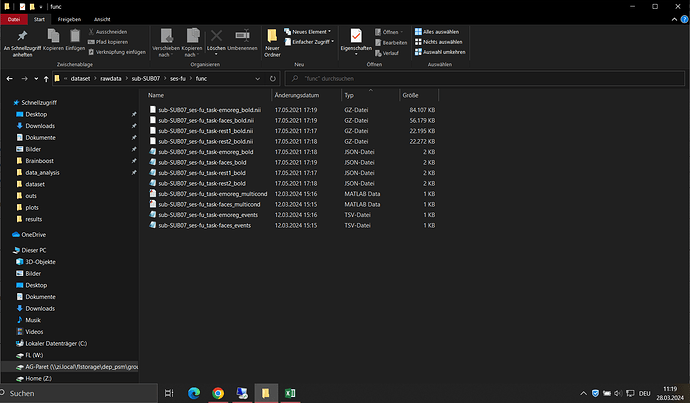
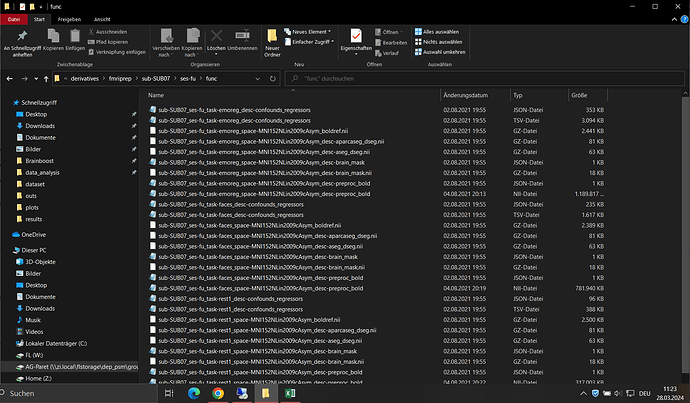
Screenshots / relevant information:
The JSON file:
{
"Name": "Faces FvS",
"BIDSModelVersion": "1.0.0",
"Input": {"subject": ["SUB07", "SUB08"], "task": ["faces"], "session": ["pre","post", "fu"]},
"Description": "A simple two-condition contrast",
"Nodes": [
{
"Level": "Run",
"Name": "run_level",
"GroupBy": ["run", "subject", "session"],
"Model": {"X": [1, "face", "scrambled"], "Type": "glm"},
"Contrasts": [
{
"Name": "FvS",
"ConditionList": ["face", "scrambled"],
"Weights": [1, -1],
"Test": "t"
}
]
},
{
"Level": "Subject",
"Name": "subject",
"GroupBy": ["contrast", "subject"],
"Model": {
"X": [1],
"Type": "meta"
},
"DummyContrasts": {"Test": "t"}
},
{
"Level": "Session",
"Name": "session_level",
"GroupBy": ["contrast", "subject", "session"],
"Model": {
"X": [1],
"Type": "meta"
},
"DummyContrasts": {"Test": "t"}
},
{
"Level": "Dataset",
"Name": "dataset_level",
"GroupBy": ["contrast", "session"],
"Model": {"X": [1], "Type": "glm"},
"DummyContrasts": {"Test": "t"}
}
]
}
Any ideas on where the problem is would be much appreciated!
Mirus