Hi,
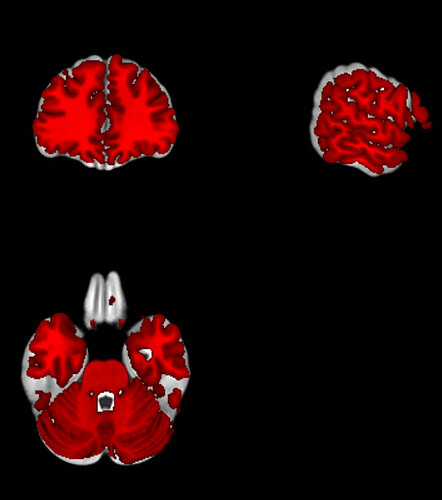
I averaged the T1 templates registered in the mni space by fmriprep for my entire sample. However I am not sure why there is a mismatch with other templates like ch2better available in MRIcron. As you can the ch2better (in red) is slightly smaller then my averaged group template. That cause me a little bit of issues so far to plot my activations on a 3D render.
Hi @simon.thibault,
Is ch2better in MNI space? Is it the same MNI space that you used for fMRIPrep (i.e., MNI152NLin6Asym vs MNI152NLin2009cAsym).
Best,
Steven
Hi Steven,
ch2better should be in the MNI space.
Indeed, it looks a bit better with MNI152NLin2009cAsym.
I was naively thinking that all templates in the MNI would be the same but it looks like it is not the case.
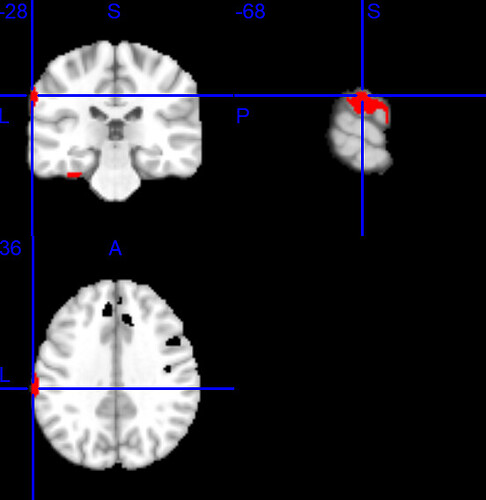
Ultimately I would like to use a render to represent my activations it looks like the fsaverage file from freesurfer is also “smaller” because I cannot see the most lateral activations. I am having the same issue in spm12 with their render (see image), where the red dot represent a cortical activation on my averaged template registered to MNI152NLin2009cAsym but this clearly out of the brain with the render (while it is not on the averaged template).
Does the freesurfer function mri_vol2surf should make the trick?
I also noticed in the anat folder of fmriprep there are surf.gii images for each subject but these images are most likely in the native space because of a mismatching number of vertices. I was one time considering avering these images but given there are not in the same space that makes thing a bit complex.
Best,
Simon
If you want a surface render of your activations, you can do your analysis in surface space (e.g., using fsaverage output space or the --cifti-output derivatives), or do your analysis in MNI space and projecting it to an fsaverage surface (e.g., with mri_vol2surf).
Depends on your command used to run fMRIPrep, in particular the output spaces. Having the full filenames would help to figure out what they are.
Best,
Steven
Here are the names of the files I was mentionning
sub-231_ses-001_rec-normalised_hemi-L_inflated.surf.gii
sub-231_ses-001_rec-normalised_hemi-L_midthickness.surf.gii
sub-231_ses-001_rec-normalised_hemi-L_pial.surf.gii
sub-231_ses-001_rec-normalised_hemi-L_smoothwm.surf.gii
Yah, those are native space.
Is there a way to move these file in MNI space and average them?
Thanks for your help!
MNI is a volumetric template, you would need a surface template (e.g., fsavergae or fsLR/cifti). Also why do you want to average them?
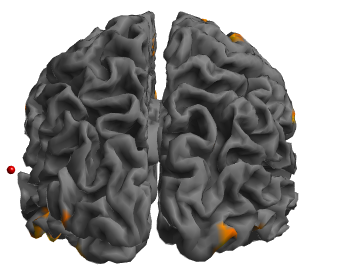
Because (but I may be wrong), but there is something weird going on with the fsaverage template (fsaverage5_lh.pial_avg.surf.gii) when I use the workbench command volume-to-surface-mapping. From what I can see in MRIcron I should have a large continuous cluster in the left fronto-parietal areas. While in workbench I do only have some scattered clusters. So I was thinking it may come from the fsaverage template and that by averaging surf.gii images created by fmriprep that would solve this issue but I am not so sure right now.
Note that by default SPM normalizes to an average sized brain, while most other tools (ANTS, FSL, etc) average to the MNI template which is larger than the average sized brain, you can see this nicely in Figure 1 of Horn et al. (2018). This is why MRIcroGL includes both the mni152 and spm152 template. Furthermore, I think it is always challenging to project statistical maps from voxels to mesh-based meshes. If you calculate values in statistics, I think you should consider voxel-based volume rendering, like MRIcroGL. If you compute your statistics based on per-vertex data, use a mesh based visualization tool like Surfice.
Ok thanks, I was thinking it would be much easier than this and that I was not able doing it just because I was not properly using different tools.
I will probably stick to MRIcroGL also because I have subcortical activations to show anyway.
Hello,
I am reopening this thread.
I was wondering, does Surfice allow to do mutlislices figure?
If not is there a way to use a color in Surfice with MRIcron or MRIcroGL or vice versa (I would like to use spectrum that is only in MRIcron or Kelvin that is only Surfice).
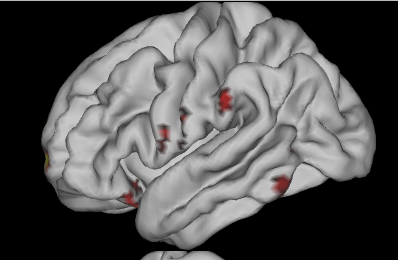
It looks like Surfice, even if the data have been estimated on a voxel-based volume, is doing quite a good job. Is that ok to overlay a voxel-based volume on a template such lh.inflated1.surf.gii? Or am I committing a huge mistake?
You can use a Python for loop and the gl.savebmp. The menu item Scripting/Templates/subcortical would give you a skeleton to achieve this.