Summary of what happened:
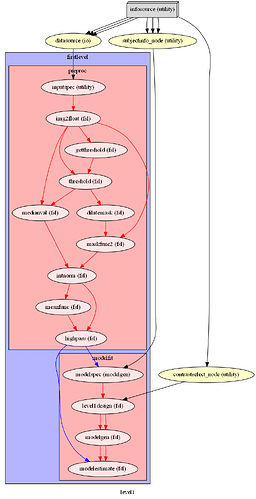
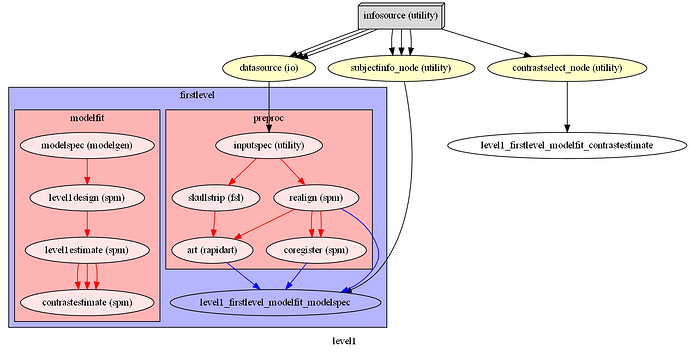
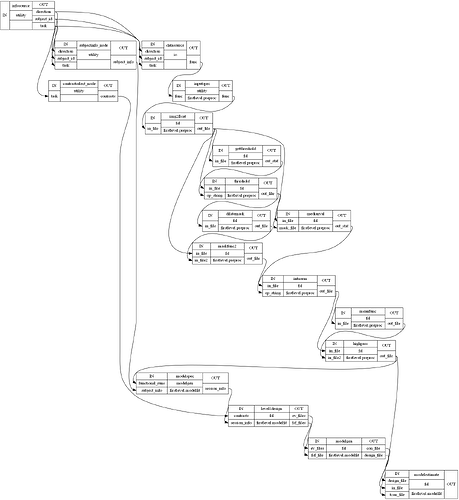
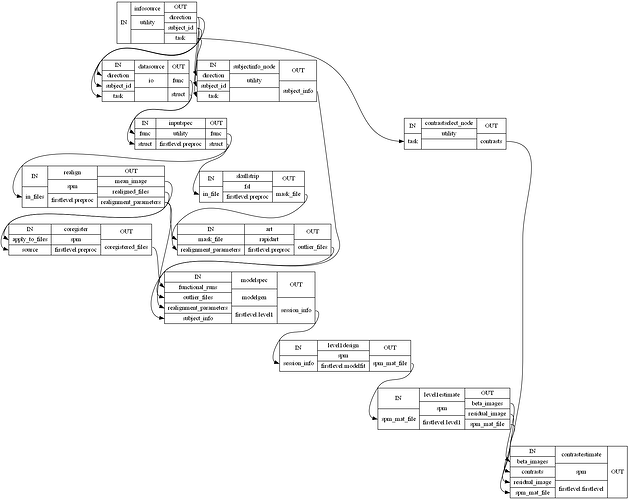
I am attempting to compare an FSL Preprocessing Pipeline with an SPM one. However, NiPype visualizations of nested workflows appear to vary when output using simple_form=True, making it confusing to determine whether nodes and edges are being defined correctly. Things however, look better when setting simple_form=False, so this may just be a bug in the visualization code?
Command used (and if a helper script was used, a link to the helper script or the command generated):
Pipeline 1 (FSL):
l1pipeline = pe.Workflow(name="level1")
l1pipeline.base_dir = os.path.abspath('./fsl/workingdir')
l1pipeline.config = {
"execution": {
"crashdump_dir": os.path.abspath('./fsl/crashdumps')
}
}
l1pipeline.connect([
(infosource, datasource, [('subject_id', 'subject_id'),
('task','task'),
('direction','direction')]),
(infosource, subjectinfo_node, [('subject_id', 'subject_id'),
('task','task'),
('direction','direction')]),
(infosource, contrastselect_node, [('task','task')]),
(subjectinfo_node, firstlevel, [
('subject_info', 'modelfit.modelspec.subject_info')]),
(datasource, firstlevel, [
('func', 'preproc.inputspec.func')]),
(contrastselect_node, firstlevel, [
('contrasts', 'modelfit.level1design.contrasts')])
])
Pipeline 2 (SPM):
l1pipeline = pe.Workflow(name="level1")
l1pipeline.base_dir = os.path.abspath('spm_tutorial/workingdir')
l1pipeline.connect([
(infosource, datasource, [('subject_id', 'subject_id'),
('task','task'),
('direction','direction')]),
(infosource, subjectinfo_node, [('subject_id', 'subject_id'),
('task','task'),
('direction','direction')]),
# Connecting contrasts from task info to contrast selection
(infosource, contrastselect_node, [('task', 'task')]),
# # Connecting functional data from datasource to preprocessing in firstlevel workflow
(datasource, firstlevel, [
('func', 'preproc.inputspec.func')]),
(datasource, firstlevel, [
('struct', 'preproc.inputspec.struct')]),
# Connect subject info to firstlevel
(subjectinfo_node, firstlevel, [('subject_info', 'modelfit.modelspec.subject_info')]),
# Connect contrasts to firstlevel design
(contrastselect_node, firstlevel, [('contrasts', 'modelfit.contrastestimate.contrasts')])
Version:
1.8.6
Environment (Docker, Singularity / Apptainer, custom installation):
Python, Jupyter Notebook within VSCode
Data formatted according to a validatable standard? Please provide the output of the validator:
N/A