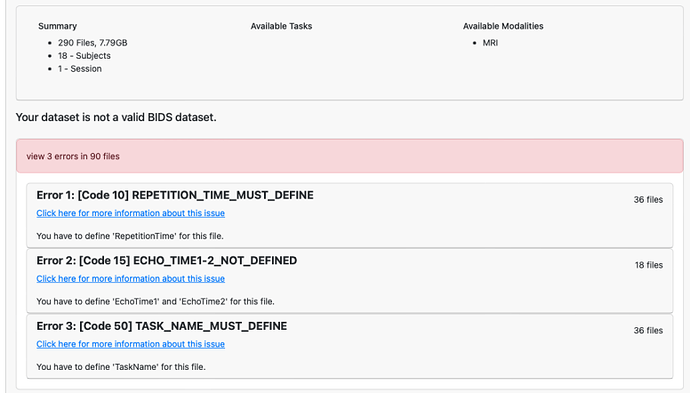
Hi there,
I am trying to upload my BIDS formatted dataset on Open Neuro (first-time user) and I encounter errors that I do not understand and cannot fix. I tried running it though BIDS validator (version 1.14.0) as recommended by Open Neuro help desk but it basically gives me the same error.
I have attached a screenshot of the BIDS validator error listing I get below (it’s identical in OpenNeuro):
It affects my func (code 10 and 50) and fmap (code 15) json files for all subjects and runs.
The issue is that these parameters it says are missing are actually there in the json files.
I looked at previous posts with similar errors and tried using a json linter to check the syntax validity but it seems my files are valid.
Here is an example of one of my json file content (bold.json):
{
"acqpar": [
{
"Modality": "MR",
"Manufacturer": "SIEMENS ",
"InstitutionName": "XXX ",
"TaskName": "Naming to definitions",
"InstitutionAddress": "XXX",
"ReferringPhysicianName": "SNAME ",
"StationName": "AWP66023",
"StudyDescription": "Brain Research_Current^SNAME",
"ProcedureCodeSequence": [
{
"CodeValue": "MRBR"
}
],
"SeriesDescription": "AP_MB4_BOLD_DEFN_B1 ",
"InstitutionalDepartmentName": "XXX ",
"PerformingPhysicianName": " ",
"OperatorsName": " ",
"ManufacturerModelName": "Prisma",
"BodyPartExamined": "BRAIN ",
"ScanningSequence": "EP",
"SequenceVariant": "SK",
"ScanOptions": "PFP\\FS",
"MRAcquisitionType": "2D",
"SequenceName": "*epfid2d1_72",
"AngioFlag": "N ",
"SliceThickness": 2.5999999046326,
"RepetitionTime": 1000,
"EchoTime": 30,
"NumberOfAverages": 1,
"ImagingFrequency": 123.241996,
"ImagedNucleus": "1H",
"EchoNumbers": 1,
"MagneticFieldStrength": 3,
"SpacingBetweenSlices": 2.6000000625534,
"NumberOfPhaseEncodingSteps": 63,
"EchoTrainLength": 63,
"PercentSampling": 100,
"PercentPhaseFieldOfView": 100,
"PixelBandwidth": 2570,
"DeviceSerialNumber": "66023 ",
"SoftwareVersions": "syngo MR E11",
"ProtocolName": "AP_MB4_BOLD_DEFN_B1 ",
"TransmitCoilName": "Body",
"AcquisitionMatrix": [72,0,0,72],
"InPlanePhaseEncodingDirection": "COL ",
"FlipAngle": 80,
"VariableFlipAngleFlag": "N ",
"SAR": 0.06474366161484001,
"PatientPosition": "HFS ",
"StudyID": "1 ",
"SeriesNumber": 11,
"AcquisitionNumber": 11,
"InstanceNumber": 11,
"ImagePositionPatient": [-639.66156442819,-692.18637022924,2.7903423194668],
"ImageOrientationPatient": [0.99699876671064,0.03325176011183,-0.0699126571296,-0.0348159321823,0.9991672722264801,-0.0212747027657],
"FrameOfReferenceUID": "1.3.12.2.1107.5.2.43.66023.1.20230317114445682.0.0.0",
"SliceLocation": -57.814503894139,
"SamplesPerPixel": 1,
"PhotometricInterpretation": "MONOCHROME2 ",
"Rows": 504,
"Columns": 504,
"PixelSpacing": [2.638888835907,2.638888835907],
"BitsAllocated": 16,
"BitsStored": 12,
"HighBit": 11,
"PixelRepresentation": 0,
"SmallestImagePixelValue": 0,
"LargestImagePixelValue": 1454,
"WindowCenter": 612,
"WindowWidth": 1261,
"WindowCenterWidthExplanation": "Algo1 ",
}
]
}
Any idea where the errors are coming from? What am I missing here?
Thank you for reading this!
Angelique